Figure 5.
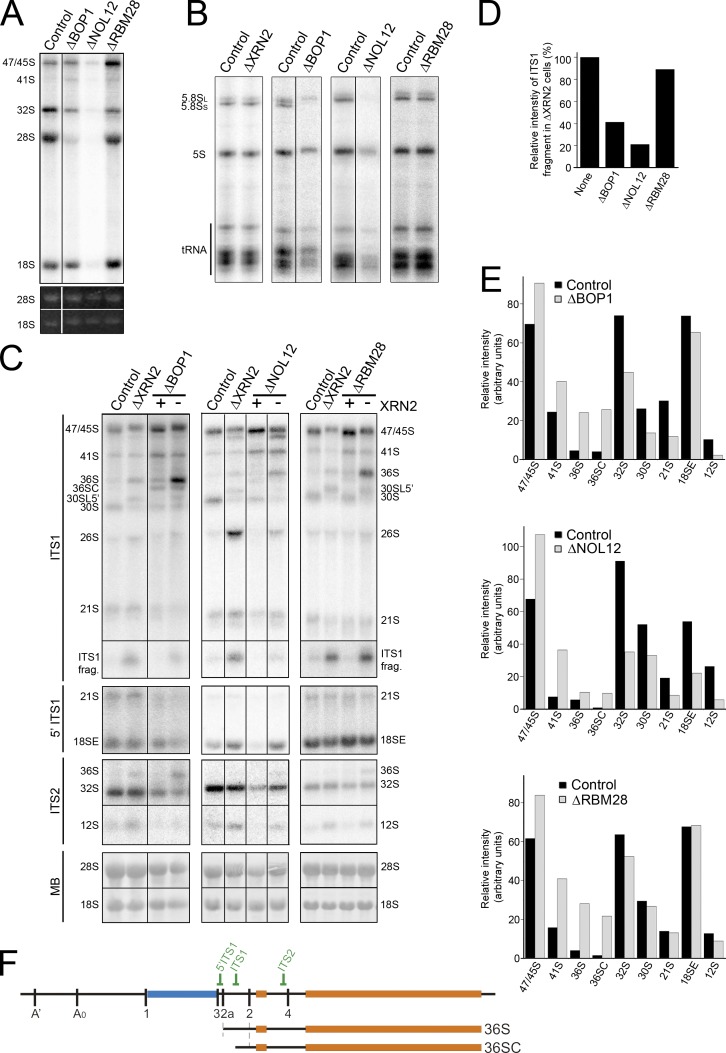
BOP1, NOL12, and RBM28 are required for site 2 cleavage in ITS1. (A and B) HeLa cells depleted of BOP1, RBM28, or NOL12 using RNAi or control cells were pulse labeled with 32P orthophosphate and then incubated with normal media for 3 h. RNA was recovered, separated using glyoxal/agarose gel electrophoresis (A) or denaturing PAGE (B), and visualized using a phosphorimager and ethidium bromide staining (UV). Positions of the mature and precursor RNAs are indicated on the left. Note that reduced tRNA production observed after depletion of BOP1 and NOL12 was not consistently seen and likely reflects the exceptionally strong effect the siRNAs had on the cells. (C) RNA extracted from control HeLa cells or HeLa cells depleted of ribosome biogenesis factors either alone or together with XRN2 (indicated at top) was analyzed by Northern blotting using probes that recognize either ITS2, ITS1, or the 5′ end of ITS1 (as indicated on the left). RNA loading was monitored by methylene blue staining of the mature 18S and 28S rRNAs (MB). In codepletion experiments, the presence (+) or depletion (−) of XRN2 is indicated at the top of the relevant lanes. (D) The relative intensity of the ITS1 fragment is plotted for the XRN2 and XRN2 double knockdowns (C), which is representative of three experiments. (E) The relative intensity of the pre-rRNA intermediates detected in C, which is representative of three experiments, are plotted (black, control; gray, knockdown). (F) Schematic representations of 47S pre-rRNA and relevant pre-rRNAs. Relative positions of Northern blot probes are indicated. Black lines in A–C indicate intervening lanes that have been removed.