Both LMNB1 depletion and overexpression inhibit proliferation, but only LMNB1 overexpression induces senescence.
Abstract
The nuclear lamina consists of A- and B-type lamins. Mutations in LMNA cause many human diseases, including progeria, a premature aging syndrome, whereas LMNB1 duplication causes adult-onset autosomal dominant leukodystrophy (ADLD). LMNB1 is reduced in cells from progeria patients, but the significance of this reduction is unclear. In this paper, we show that LMNB1 protein levels decline in senescent human dermal fibroblasts and keratinocytes, mediated by reduced transcription and inhibition of LMNB1 messenger ribonucleic acid (RNA) translation by miRNA-23a. This reduction is also observed in chronologically aged human skin tissue. To determine whether altered LMNB1 levels cause senescence, we either increased or reduced LMNB1. Both LMNB1 depletion and overexpression inhibited proliferation, but only LMNB1 overexpression induced senescence, which was prevented by telomerase expression or inactivation of p53. This phenotype was exacerbated by a simultaneous reduction of LMNA/C. Our results demonstrate that altering LMNB1 levels inhibits proliferation and are relevant to understanding the molecular pathology of ADLD.
Introduction
The nuclear lamina underlies the inner nuclear membrane and consists of a meshwork of intermediate filament proteins: the A- and B-type lamins. B-type lamins (lamins B1 and B2) are ubiquitously expressed in all cell types, whereas expression of LMNA/C (lamin A/C) is largely restricted to somatic cells (Stewart and Burke, 1987; Röber et al., 1989). Lamins provide a scaffold for a variety of nuclear proteins and maintain the architectural integrity of interphase nuclei. Mutations in the LMNA gene are associated with over a dozen diseases, collectively called laminopathies (Burke and Stewart, 2006). Laminopathies affect skeletal homeostasis, muscle, heart, and vascular tissues and cause the accelerated aging syndromes Hutchinson-Gilford progeria syndrome (HGPS) and atypical Werner syndrome (Chen et al., 2003; De Sandre-Giovannoli et al., 2003; Csoka et al., 2004; Eriksson et al., 2003).
B-type lamins have been implicated in regulating DNA replication (Moir et al., 1994), RNA synthesis (Tang et al., 2008), induction of the oxidative stress response (Malhas et al., 2009), mitotic spindle assembly (Tsai et al., 2006), and the spatial distribution of chromosomes (Guelen et al., 2008). To date, no loss-of-function or dominant-acting missense mutations of B-type lamins have been identified. A possible explanation for this is that loss of B-type lamins, as in mice, results in perinatal death, with defects in the lungs, skeleton, neuronal migration, and central nervous system (CNS; Vergnes et al., 2004; Burke and Stewart, 2006; Worman et al., 2010; Coffinier et al., 2011; Kim et al., 2011). In contrast, duplication of the LMNB1 locus, resulting in increased LMNB1 (lamin B1) expression, is associated with adult-onset autosomal dominant leukodystrophy (ADLD), a disease affecting myelination of the CNS with severe neurological defects (Padiath and Fu, 2010). LMNB1 is also increased in lymphoblasts and fibroblasts from ataxia telangiectasia (AT) patients, another disease associated with neurological defects (Barascu et al., 2012). However, mechanistic insights into how LMNB1 overexpression damages cells or why the brain and CNS are particularly susceptible to fluctuations of LMNB1 remain elusive.
Several recent studies have highlighted the importance of LMNB1 in regulating proliferation and senescence of cultured human cells (Shimi et al., 2011; Barascu et al., 2012; Freund et al., 2012). LMNB1 is reduced in HGPS cells and declines in normal fibroblasts as they enter replicative senescence (Scaffidi and Misteli, 2005; Taimen et al., 2009; Shimi et al., 2011; Zhang et al., 2011; Freund et al., 2012). Shimi et al. (2011) reported that LMNB1 reduction triggered senescence, whereas its overexpression delayed senescence. In contrast, Barascu et al. (2012) showed that LMNB1 overexpression causes senescence. Here, we clarify and extend these findings and provide mechanistic insight into how LMNB1 overexpression results in senescence.
We show that LMNB1 and LAP2 (lamina-associated polypeptide 2 or LEMD4) both decline in senescent primary human dermal fibroblasts and keratinocytes in vitro. We demonstrate that a reduction of LMNB1 and LAP2 also occurs during chronological aging of human skin keratinocytes in vivo. These results indicate that the nuclear lamina changes profoundly as cells enter replicative senescence, both in vitro and in vivo.
To investigate whether LMNB1 reduction is a cause or a consequence of senescence, LMNB1 was experimentally increased or decreased in primary human fibroblasts. We find that LMNB1 reduction impairs proliferation but, under normal culture conditions, does not result in senescence. In contrast, LMNB1 overexpression impairs proliferation and culminates in cellular senescence, with these effects being rescued by telomerase or inactivation of p53. Lastly, we show that cells with low levels of LMNA/C are significantly more sensitive to LMNB1 overexpression: these cells exhibit impaired proliferation, increased DNA damage at the telomeres, and senesce prematurely. These results may provide an explanation as to why ADLD manifests itself mainly in the brain, where LMNA/C levels are reduced (Jung et al., 2012).
Collectively, we show that a reduction in LMNB1 can serve as a marker for cellular aging in human skin and that stoichiometric changes in LMNB1 have significant, yet diverse, effects on cellular proliferation. These results provide mechanistic insights into how LMNB1 duplication may affect cellular function in ADLD and other diseases characterized by altered amounts of LMNB1, such as AT.
Results
Reduced LMNB1 is a characteristic of HGPS fibroblasts, normal fibroblasts undergoing replicative senescence, and fibroblasts with telomere-specific DNA damage
Progeric patient-derived fibroblasts exhibit a decrease in LMNB1 expression (Scaffidi and Misteli, 2005; Liu et al., 2011; Zhang et al., 2011). In particular, a patient-derived cell line carrying an N-terminal E145K mutation in the LMNA gene exhibited severe nuclear abnormalities (Fig. S1, a and b) and reduced levels of LMNA and LMNB1 (Fig. S1 c; Taimen et al., 2009).
Progeric cell lines, including the LMNA E145K, have shortened telomeres and undergo premature replicative senescence compared with age-matched controls (unpublished data; Bridger and Kill, 2004; Decker et al., 2009). To test whether the reduction of LMNB1 was a consequence of replicative senescence, we passaged primary human dermal fibroblasts until they entered senescence after ∼55–60 population doublings (PDs). As shown by quantitative Western blotting, LMNB1 decreased with passage to ∼10% of their original level as the cells entered senescence (Fig. S1, d and e, left). In contrast, LMNB1 did not decrease during continuous culture of fibroblasts immortalized by human telomerase reverse transcription (TERT; hTERT; Fig. S1, d and e, right). Replicative senescence was assessed by expression of senescence-associated β-galactosidase (SA-β-gal) activity (Dimri et al., 1995) and persistent DNA damage foci (d’Adda di Fagagna et al., 2003). At the start, ∼10% of primary, and ∼3% of hTERT-immortalized cells stained positive for SA-β-gal activity. After 55 PDs, ∼85–90% of primary and ∼3% of hTERT-immortalized cells stained positive for SA-β-gal (Fig. S1 f). Immunofluorescence staining for the DNA damage factor 53BP1 showed increased numbers of DNA damage foci in senescent cells compared with early passage primary or hTERT-immortalized cells (Fig. S1, g and h).
We also elicited senescence in fibroblasts by inducing telomere-specific DNA damage using a dominant-negative allele of TRF2, TRF2ΔBΔM. TRF2 is a component of the shelterin complex that protects telomeres (de Lange, 2005). Removal of endogenous TRF2 from the telomeres (by expression of TRF2ΔBΔM) triggers a DNA damage response, resulting in senescence (Karlseder et al., 1999). As expected, expression of TRF2ΔBΔM impaired proliferation and was accompanied by a reduction in LMNB1 (Fig. S1 i). Together, these results demonstrate that LMNB1 loss is a characteristic of senescent cells.
Next, we investigated whether other components of the nuclear lamina were reduced during senescence. The LAP2 exists as six alternatively spliced isoforms, four of which (LAP2-β, -γ, -δ, and -ε) interact with LMNB1 and localize to the inner nuclear membrane (Dorner et al., 2006). LAP2-α has been implicated in regulating cell proliferation in murine and human cells (Pekovic et al., 2007; Naetar et al., 2008). By Western blot analysis and immunofluorescence staining, using an antibody recognizing all LAP2 isoforms (Fig. S2, a and b), we observed that most isoforms were reduced in senescent fibroblasts (Fig. S2 a). We confirmed these results using an antibody specific to the LAP2-α isoform (Fig. S2 a). To investigate whether the reduction of LMNB1 and LAP2-α was specific to senescent cells, we arrested normal dermal fibroblasts by serum starvation or by growing them to confluence. In agreement with previous results (Pekovic et al., 2007; Naetar et al., 2008), LAP2-α decreased in response to serum starvation–induced cell cycle exit (Fig. S2 c), whereas LMNB1 did not (Fig. S2 c). Similarly, cells driven into quiescence by growth to complete confluence (as verified by Ki-67 staining) expressed lower levels of LAP2-α (Fig. S2 d). In conclusion, although LMNB1 levels decrease specifically in senescent cells, LAP2-α declines in cells exiting the cell cycle, regardless of whether growth arrest was induced by confluence, serum starvation, or replicative senescence. Furthermore, these results reveal that the nuclear lamina is significantly altered during cellular senescence.
LMNB1 reduction is associated with senescence in human keratinocytes
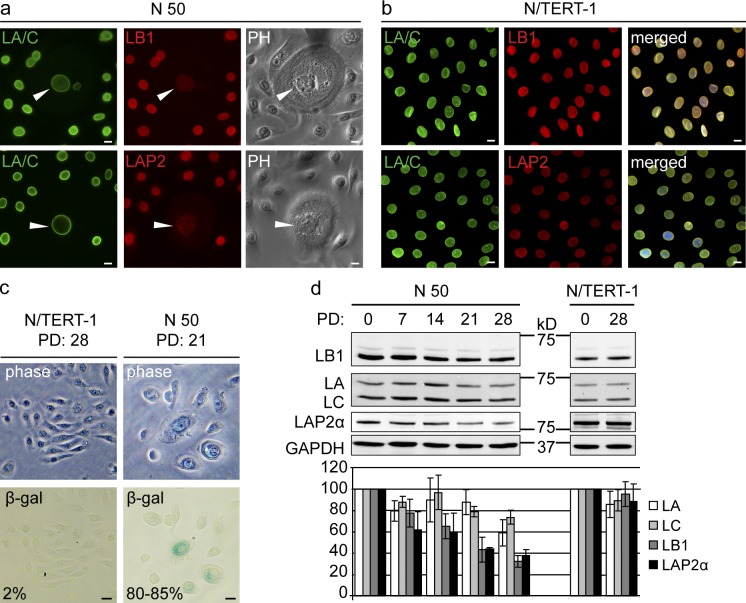
The reduction in LMNB1 in senescent fibroblasts led us to investigate whether LMNB1 reduction is a marker for senescence in keratinocytes, the major cell type found in skin. We cultured two primary keratinocyte lines (N 49 and N 50) until they reached replicative senescence (after ∼28 PDs) and compared them with hTERT-immortalized keratinocytes (N/TERT-1; Dickson et al., 2000). Initially, primary keratinocytes were nearly indistinguishable from their N/TERT-1 counterparts: they grew as distinct clusters and stained strongly for LMNA/C, LMNB1, and LAP2 (Fig. 1, a and b). During subsequent culture of N 49 and N 50, large cells with a flattened morphology appeared, whereas N/TERT-1 cells retained a normal morphology. The number of flattened cells increased with successive PDs (Fig. 1 a). The flattened cells showed decreased staining for LMNB1 and LAP2 (Fig. 1 a, arrowheads), and after 21 PDs, ∼85–90% of cells stained positive for SA-β-gal activity (Fig. 1 c). The reduction in LMNB1 and LAP2-α was confirmed by quantitative Western blotting: after 28 PDs, LMNB1 and LAP2-α decreased to ∼30 and 40%, respective to their original levels (Fig. 1 d). No change in LMNB1 was observed in two independently maintained N/TERT-1 cell lines (Fig. 1 d). These observations parallel our results from senescent dermal fibroblasts and demonstrate that a reduction in LMNB1 and LAP2-α marks keratinocyte senescence in vitro.
Figure 1.
Loss of LMNB1 in senescent human keratinocytes. (a and b) Immunofluorescence staining of primary keratinocytes (N 50; a) and hTERT-positive keratinocytes (N/TERT-1; b) with LMNA/C (LA/C), LMNB1 (LB1), and LAP2 antibodies. Merged images are shown for N/TERT-1 cells only. (right) Phase contrast (PH) images are shown for N 50. Arrowheads show large flattened cells. Bars, 10 µm. (c) SA-β-gal staining of hTERT-positive (N/TERT-1) and primary (N 50) keratinocytes at PD 28 and 21, respectively. Same fields shown in phase contrast (top) and bright field (bottom). The percentages of SA-β-gal–positive cells are indicated. Bars, 20 µm. (d) LMNB1, LMNA/C, and LAP2-α levels during a 28 PD time course of primary N 50 cells (left) and one independently passaged N/TERT-1 line (right) by Western blotting. Quantified signal intensities of LMNB1, LMNA/C, and LAP2-α and normalized to GAPDH control are plotted below (n = 3). The results are presented as the means ± SD of three independent experiments.
Reduced levels of LMNB1 and LAP2 are associated with normal skin aging and terminal keratinocyte differentiation in vivo
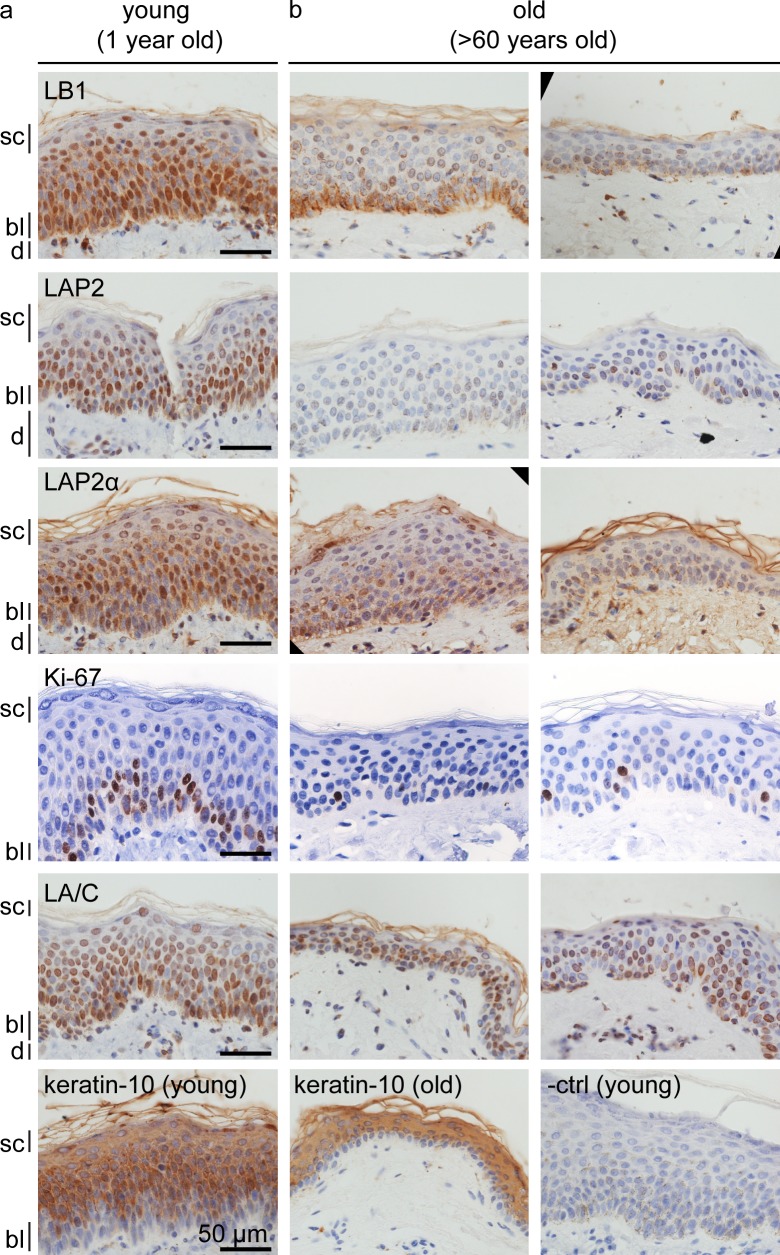
Because senescent human fibroblasts and keratinocytes show a reduction of LMNB1, we investigated whether these findings are relevant to normal human skin aging in vivo. We analyzed LMNB1, LAP2, LAP2-α, Ki-67, LMNA/C, and keratin-10 expression in skin sections from three different normal donors (aged 60 and above) and one donor (aged 1). Immunohistochemical staining revealed robust levels of LMNB1, LAP2, LAP2-α, and LMNA/C in the majority of young epidermal keratinocytes (Fig. 2 a). In contrast, in skin from old donors, LMNB1, LAP2, and LAP2-α were reduced and mostly confined to keratinocytes residing in the basal layer of the epidermis (Fig. 2 b), whereas LMNA/C or keratin-10 staining was not affected by donor age. Furthermore, the number of Ki-67–positive cells in the basal layer was dramatically reduced in aged skin. Keratinocytes migrate from the basal layer to the stratum corneum, where they terminally differentiate, arrest in G1, and ultimately lose their nuclei to become corneocytes. LMNB1 and LAP2 decreased during these terminal stages of differentiation, even in young skin (Fig. 2 a, stratum corneum). Collectively, our results show that a reduction in LMNB1 and LAP2 marks senescence of primary human fibroblasts and keratinocytes in vitro as well as during normal aging of human skin in vivo.
Figure 2.
Loss of LMNB1 and LAP2 during normal skin aging and terminal keratinocyte differentiation in vivo. (a and b) Immunohistochemistry of paraffin sections of normal human skin from young (1 yr; a) versus old (>60 yr; b) individuals. Antibodies are LMNB1 (LB1), LAP2, LAP2-α, LMNA/C (LA/C), Ki-67, keratin-10, and negative control (−ctrl). Different skin layers are indicated: stratum corneum (sc), basal layer (bl), and dermis (d).
Senescence-associated LMNB1 decline is regulated at the mRNA level and by miR-23a
The correlation of LMNB1 reduction with senescence in vitro and in vivo prompted us to investigate how LMNB1 is regulated. First, we measured LMNB1 and LMNA/C mRNA levels by quantitative RT-PCR (qRT-PCR) in normal versus senescent fibroblasts. As shown in Fig. S3 a, in contrast to LMNA/C, which remains stable, LMNB1 transcripts decrease ∼20-fold in senescent cells, revealing that reduced LMNB1 mRNA correlates with the decline in LMNB1 protein.
miRNAs regulate protein expression by binding to mRNAs and targeting them for degradation or by inhibiting translation (Huntzinger and Izaurralde, 2011). A previous study showed that miR-23a regulates Lmnb1 during postnatal CNS development in mice (Lin and Fu, 2009). The mouse Lmnb1 3′UTR contains three miR-23a binding sites that are conserved between murine and human LMNB1 genes. We investigated whether miR-23a may regulate LMNB1 in human cells and found that miR-23 increases ∼2.5-fold in senescent cells (Fig. S3 b). We then fused the wild-type LMNB1 3′UTR, or a LMNB1 3′UTR containing point mutations in the putative miR-23a binding sites (LMNB1 3′UTR mutant; Fig. S3 c), to a luciferase reporter and cotransfected the reporter construct with miR-23a and a nonspecific control, miR-882, into HeLa cells. Transfection of miR-23a decreased luciferase activity (Fig. S3 d), whereas transfection of miR-822 had no effect. Luciferase levels were restored after the introduction of point mutations into the miR-23a binding sites, demonstrating that miR-23a specifically targets the LMNB1 3′UTR. Next, we determined whether expression of miR-23a in fibroblasts reduced LMNB1 mRNA and/or protein. As shown in Fig. S3 e, LMNB1 mRNA levels were not significantly reduced by miR-23a or miR-822 transfection (Fig. S3 e). However, LMNB1 protein decreased ∼30% by miR-23a, suggesting that miR-23a blocks translation (Fig. S3 f). These results suggest that miR-23a is involved in the regulation of LMNB1 in human cells and may contribute to the decrease in LMNB1 protein during cellular senescence.
LMNB1 decline is a hallmark and a consequence of senescence but does not cause senescence
The senescence-associated loss of LMNB1 raises the important question whether LMNB1 reduction is a cause or a consequence of senescence. It was previously reported that inhibition of LMNB1 in WI-38 cells resulted in senescence that was rescued by inhibition of p53 or by culturing cells under hypoxic conditions (Harborth et al., 2001; Shimi et al., 2011). Furthermore, overexpression of LMNB1 in WI-38 cells increased their proliferation and delayed the onset of senescence (Shimi et al., 2011). In contrast, a recent study indicated that LMNB1 overexpression induced senescence in fibroblasts (Barascu et al., 2012).
Given these contradictory findings, we investigated the consequences of LMNB1 depletion and overexpression in human fibroblasts. To deplete LMNB1, we expressed two different shRNAs using constitutive or doxycyclin-inducible lentiviral vectors (pGIPZ and pTRIPZ, respectively). To overexpress LMNB1, we used constitutive (pBABE based) and doxycyclin-inducible (pTRIPZ based) expression vectors. All experiments were performed in two primary fibroblast lines and their hTERT-immortalized counterparts as well as in WI-38.
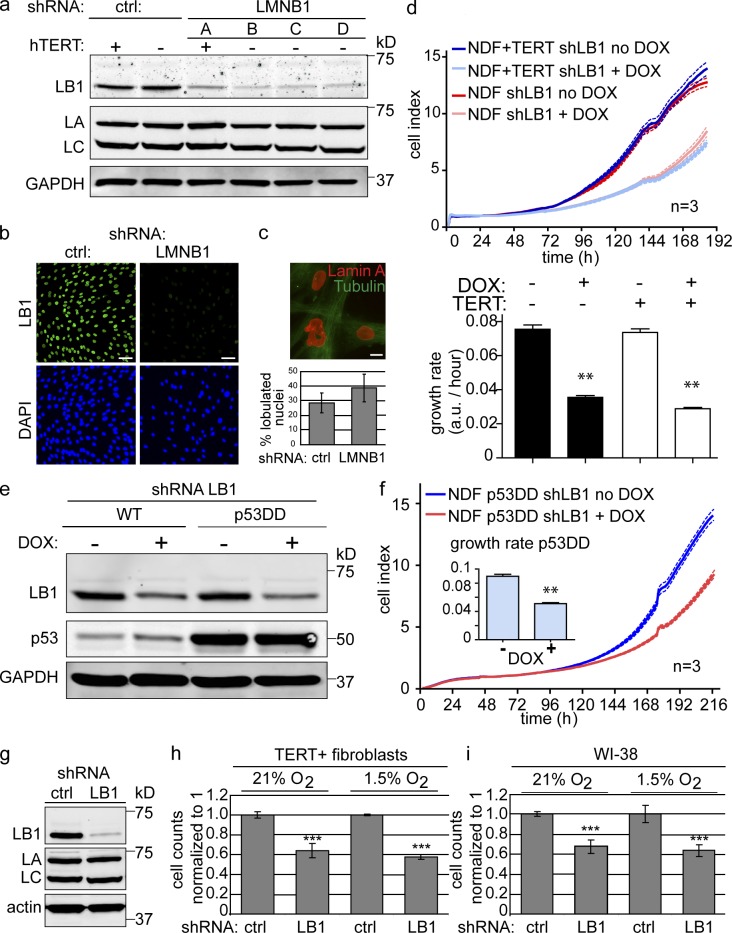
Quantitative Western blotting and immunostaining revealed a ∼90% reduction in LMNB1 after expression of LMNB1 shRNAs compared with cells expressing a scrambled control shRNA (Fig. 3, a and b). LMNB1-depleted cells exhibited increased nuclear blebbing compared with controls (28.6 ± 6.8% vs. 38.9 ± 9.53%, respectively; Fig. 3 c). We then determined the consequences of LMNB1 reduction on cell proliferation. Both hTERT-positive and hTERT-negative fibroblasts expressing the LMNB1 shRNA exhibited impaired proliferation (Fig. 3 d, top [growth curve] and bottom [growth rate]). Cells expressing the scrambled shRNA or two control cell lines treated with doxycyclin grew normally. In contrast, a reduction in LMNB2 (lamin B2), using two different shRNAs (B2a and B2b), did not result in any significant growth retardation (Fig. S4, a and b). We conclude that experimentally lowering LMNB1, but not LMNB2, has a specific inhibitory effect on cell proliferation.
Figure 3.
shRNA-mediated depletion of LMNB1 impairs proliferation but does not cause cellular senescence. (a) Western blot depicting LMNB1, LMNA/C, and GAPDH levels in clones expressing a scrambled control (ctrl) or LMNB1 shRNA (clones A, B, C, and D). (b) Immunofluorescence staining of control and LMNB1 (LB1)-depleted cells using an antibody against LMNB1 (green). DAPI staining is in blue. Bars, 50 µm. (c) Nuclear abnormalities in LMNB1-deficient fibroblasts. Bar, 10 µm. Quantification of nuclear abnormalities is shown below. The results are presented as the means ± SD of three independent experiments. (d) LMNB1-deficient fibroblasts exhibit growth retardation. (top) Growth curve of primary and hTERT-positive normal dermal fibroblasts (NDFs) stably infected with doxycyclin (DOX)-inducible LMNB1 shRNA construct. Doxycyclin induced (LMNB1 deficient: light blue and red) and noninduced (dark blue and dark red) are shown. Dotted lines indicate SEM (n = 3). (bottom) 6-d mean growth rate of doxycyclin induced (+) versus noninduced (−). Error bars represent ± SEM. **, P < 0.01. (e) Expression of dominant-negative p53 (p53DD) does not rescue LMNB1 loss–induced growth retardation. Doxycyclin-inducible down-regulation of LMNB1 in cells expressing vector control (first and second lanes) or a dominant-negative allele of p53 (p53DD; third and fourth lanes). (f) Growth curve of p53DD-expressing fibroblasts ± doxycyclin-inducible LMNB1 shRNA (dotted lines indicate SEM [n = 3]). Inset shows 8-d mean growth rate for noninduced (−) and induced (+). Error bars indicate ± SEM. **, P < 0.01. (g) LMNB1 loss–induced proliferation defect is not rescued by growth under low oxygen (hypoxia 1.5% O2) conditions. Western blot of cells constitutively expressing a scrambled control shRNA or an shRNA against LMNB1. LA, LMNA; LC, LMNC. (h) Proliferation assay of hTERT-positive dermal fibroblasts cells expressing scrambled control (sc) or shRNA against LMNB1 under normal (21% O2) and hypoxic (1.5% O2) conditions. (i) Proliferation assay of WI-38 cells expressing control or LMNB1 shRNA. Assays were performed in triplicates, and means ± SD from three independent experiments are shown. ***, P < 0.001. Cell numbers of scrambled controls were normalized to 1. a.u., arbitrary unit; WT, wild type.
To investigate whether the reduced proliferation of LMNB1-deficient cells was caused by cells entering premature senescence, we performed the following experiments: First, a hallmark of senescence is the activation of the tumor suppressor p53. p53 inactivation, by expression of a dominant-negative allele (p53DD) that disrupts p53 tetramerization, prevents replicative senescence (Shaulian et al., 1992; Hahn et al., 2002). To investigate whether p53 inactivation rescues the proliferation defect of LMNB1-depleted cells, we reduced LMNB1 (using a doxycyclin-inducible shRNA) in cells expressing p53DD (Fig. 3 e). Consistently, p53DD did not rescue the proliferation defects induced by LMNB1 depletion, demonstrating that LMNB1-dependent growth retardation is independent of p53 (Fig. 3 f). Second, we investigated whether growth under hypoxic conditions would restore proliferation of LMNB1-depleted cells (Shimi et al., 2011). We performed proliferation assays on two different hTERT-negative and hTERT-positive fibroblast lines and WI-38, (Fig. 3 g) under normoxic (21% O2) and two hypoxic conditions (1.5 and 3.5% O2). Growth under hypoxic conditions did not rescue the LMNB1 depletion–dependent proliferation defect in any of the cell lines tested (Fig. 3, h and i). Third, we stained fibroblasts expressing either LMNB1 or a scrambled control shRNA for SA-β-gal activity. We did not detect any increase in SA-β-gal–positive cell numbers upon LMNB1 depletion (a total of 790 TERT-negative cells and 989 TERT-positive cells were counted). The discrepancy between these and previously reported results (Shimi et al., 2011) led us to investigate whether different experimental procedures may have affected the outcome of our experiment. A previous study used retroviral transduction and 3 µg/ml puromycin (puro) selection to derive cells stably expressing LMNB1 shRNA (Shimi et al., 2011). In contrast, we used lentiviral vectors expressing the LMNB1 shRNA either constitutively or under the control of a doxycyclin-inducible promoter. As judged by FACS analysis, our lentiviral transduction efficiency was high (∼90%; Fig. S5 a). Consequently, we did not clonally expand cells expressing the vector but started with a stably infected cell population. The LMNB1 levels in the infected cells were then reduced either constitutively or after doxycyclin addition. To investigate whether the reported LMNB1 loss–induced senescence may be a result of stress during clonal selection in conjunction with LMNB1 deficiency, we seeded control and LMNB1-depleted cells at different densities: sparse (10,000 cells/9.6-cm2 well) versus subconfluent (40,000 cells/9.6-cm2 well). We observed increased SA-β-gal–positive cell numbers in sparsely plated LMNB1-depleted cells compared with controls (Fig. S5, b and c). Strikingly, the same cells grown at subconfluent density did not exhibit increased SA-β-gal staining. These results suggest that although a reduction of LMNB1 results in decreased cell proliferation, this reduction does not trigger premature senescence in either primary or TERT-positive fibroblasts, unless they are subjected to additional stress, such as low density growth or high drug selection. Next, we performed FACS analysis of LMNB1-depleted versus control cells and consistently observed a slight increase in the percentage of cells in G2/M (Fig. S5 d). We also observed additional peaks to the right of the G2/M peak, suggesting chromosomal instability in these cells. However, these events were rare, and we failed to detect them by analysis of metaphase spreads.
Increased LMNB1 induces cellular senescence in human fibroblasts
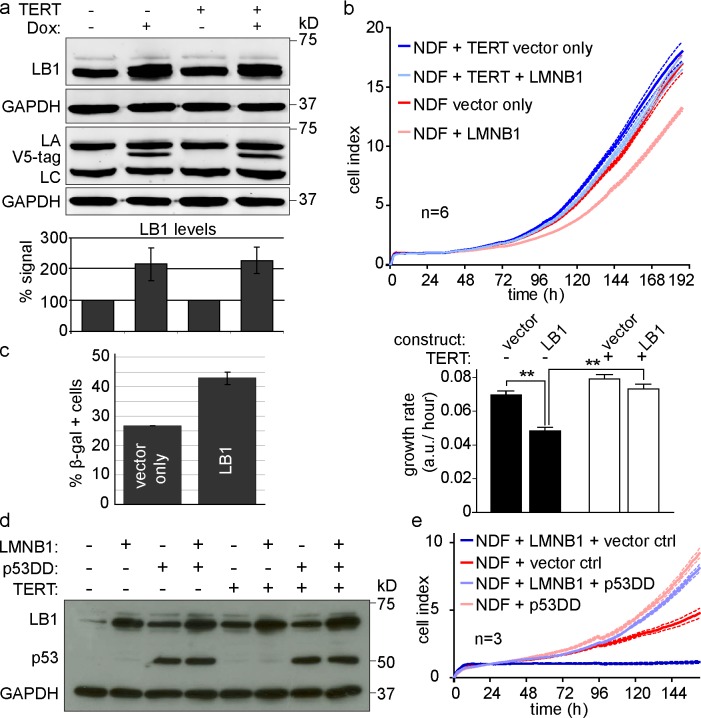
ADLD is a disease that manifests itself by widespread myelin loss in the CNS (Padiath et al., 2006). It is caused by duplication of the LMNB1 locus, resulting in increased LMNB1 protein expression. We used both retroviral- and doxycyclin-inducible lentiviral vectors to increase LMNB1 levels by two to threefold, thereby mimicking the increase of LMNB1 in ADLD (Fig. 4 a and Fig. 5 a; Padiath et al., 2006). We then investigated the consequences of increased LMNB1 on the growth of hTERT-negative and hTERT-positive cells. Increased LMNB1 led to impaired proliferation of primary fibroblasts, which was rescued by expression of hTERT (but not catalytically inactive hTERT; Fig. 4 b). Consistent with Barascu et al. (2012), the LMNB1 overexpression–induced proliferation defect led to increased numbers of SA-β-gal–positive cells (26.6% in control vs. 42.8% in LMNB1-expressing cells; Fig. 4 c). The majority of senescence-inducing signals, such as persistent DNA damage, activate the p53 pathway (Campisi and d’Adda di Fagagna, 2007). To investigate whether increased LMNB1-induced senescence could be bypassed by blocking p53, we introduced the p53DD dominant-negative allele (described in the previous paragraph) or a vector only control (pBABE-hygromycin [hygro]) into cells overexpressing LMNB1. We verified expression of each construct by Western analysis (Fig. 4 d) and determined their proliferation rate. Although we obtained hTERT-deficient clones overexpressing LMNB1 and the control vector pBABE-hygro, these cells proliferated very poorly (Fig. 4 e, dark blue line). As expected, the same cell line with normal LMNB1 levels grew significantly better (Fig. 4 e, red line). In contrast, cells expressing p53DD, regardless of whether they overexpressed LMNB1 or not, bypassed senescence and grew at roughly identical rates (Fig. 4 e, compare light blue with light red line). These results demonstrate that LMNB1 overexpression–induced senescence is rescued by inhibiting p53 or by expressing hTERT, suggesting that LMNB1 overexpression may lead to telomere-specific DNA damage.
Figure 4.
Overexpression of LMNB1 causes senescence, which is reversible by telomerase or p53 inactivation. (a) Doxycyclin-inducible overexpression of v5-tagged LMNB1 (LB1). (top) Western blot showing LMNB1 levels upon doxycyclin induction (+) in hTERT-negative and hTERT-positive normal dermal fibroblasts. Samples were run in parallel on two gels and probed with the following antibodies: LMNB1, LMNA/C (LA and LC), and v5 tag. GAPDH loading control is shown for both gels. (bottom) Quantification of LMNB1 in noninduced versus induced cells (normalized to GAPDH/hTERT negative or hTERT positive/no doxycycline [Dox]; n = 5). The means ± SD of five independent experiments are shown. (b, top) Growth curve of hTERT-negative (red and light red) and hTERT-positive (blue and light blue) cells expressing LMNB1 or a vector control. Dotted lines indicate SEM (n = 6). (bottom) Mean growth rate of cells ± LMNB1. Error bars indicate ± SEM. **, P < 0.01. (c) Quantification of SA-β-gal–positive cells in LMNB1-overexpressing cells versus vector control (n = 3). Data are presented as means ± SD from three independent experiments. (d) Expression of LMNB1 or vector control, dominant-negative p53 (p53DD), or pBABE-hygro control in hTERT-negative and hTERT-positive fibroblasts. Antibodies are p53, LMNB1, and GAPDH. (e) Growth curve of hTERT-deficient fibroblasts overexpressing LMNB1 (or pBABE-neo) in the presence of dominant-negative p53DD or pBABE-hygro control (ctrl). Dotted lines indicate SEM (n = 3). a.u., arbitrary unit; NDF, normal dermal fibroblast.
Figure 5.
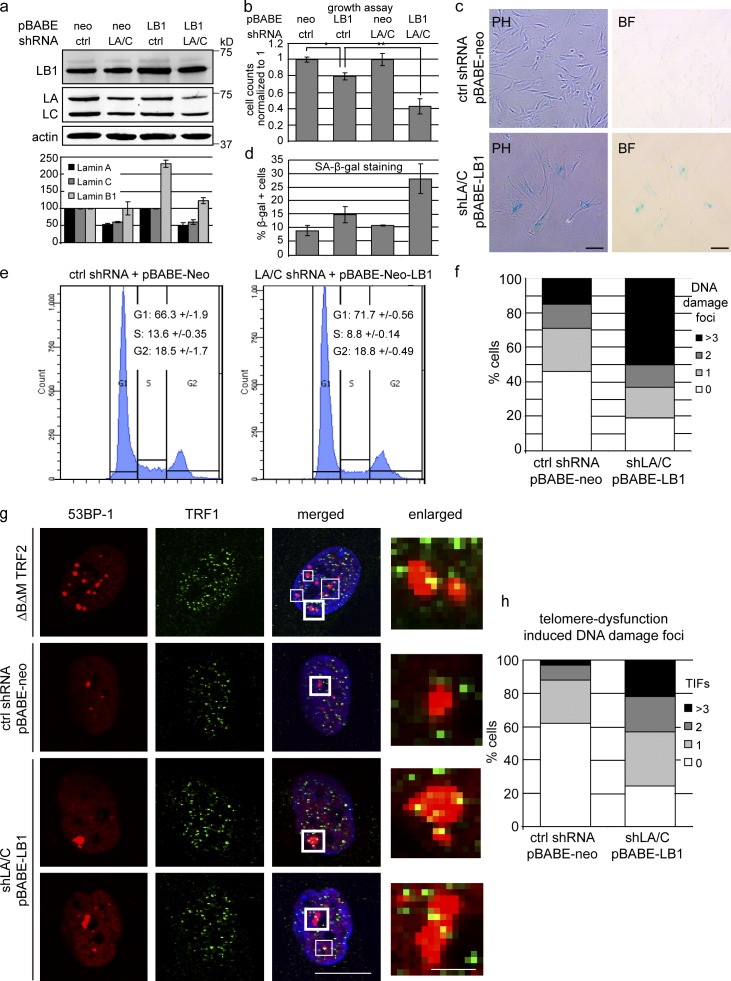
Reduced levels of LMNA/C exacerbate the effects of LMMB1 overexpression. (a, top) Western blot showing LMNB1 (LB1) and LMNA/C (LA/C) levels in cells expressing pBABE-neo control or pBABE-neo-LMNB1 in cells previously transduced with control (ctrl) or LMNA/C shRNA. Antibodies are LMNB1, LMNA/C, and actin. (bottom) Quantification of LMNB1 and LMNA/C levels normalized to actin (n = 3). Data are from three independent experiments. (b) Proliferation assay of cells expressing pBABE-neo or pBABE-neo-LMNB1 in cells expressing control shRNA (first two bars) or in cells expressing shRNA against LMNA/C (last two bars). *, P < 0.05; **, P < 0.01; n = 4. (c) SA-β-gal staining of control versus LMNB1 in shLMNA/C cells (PH, phase contrast; BF, bright field). Bars, 50 µm. (d) Quantification of SA-β-gal–positive cells in cells described in b. (n = 4). (e) Cell cycle analysis of control versus LMNB1 in shLMNA/C cells. Percentages of cells in different cell cycle stages are indicated (n = 4). (f) Quantification of DNA damage foci by 53BP1 staining in control versus LMNB1 in shLMNA/C cells. At least 400 cells were counted for each condition. (g) Confocal immunofluorescence microscopy showing telomere dysfunction–induced DNA damage foci (TIFs). Cells were stained with 53BP-1 (red) and TRF1 (green). Shown are cells expressing TRF2ΔBΔM (positive control) or vector control (scrambled shRNA + pBABE-neo) and cells with reduced levels of LMNA/C overexpressing LMNB1 (bottom two rows). Merged images also show DAPI staining. Bar, 20 µm. White boxes highlight TIFs. Thick framed white boxes are enlarged on the right. Bar, 2.5 µm. (h) Quantification of TIFs from control versus shLMNA/C:LMNB1 cells. Percentages of cells showing zero, one, two or more than three TIFs are shown. 100 cells were counted for each condition. Error bars represent ± SD.
Reduced levels of LMNA/C exacerbate LMNB1 overexpression–induced phenotypes
ADLD affects mainly the CNS. Likewise, Lmnb1 and b2 double-null mouse embryos display developmental defects, especially in the CNS (Coffinier et al., 2011; Kim et al., 2011). Why is the brain more susceptible to fluctuation of B-type lamins than other tissues? One difference is that neurons express lower levels of LMNA/C than, for instance, tissues of mesenchymal origin (Zhang et al., 2011). To test whether a reduction in LMNA/C renders cells more susceptible to alterations in LMNB1, we increased LMNB1 in cells with reduced LMNA/C. Quantitative Western blotting revealed that LMNA/C was reduced by 40–50% (Fig. 5 a), yet these cells proliferated normally. We then increased LMNB1 in these and control fibroblasts. Quantitative Western analysis showed that LMNB1 levels had doubled in control cells (Fig. 5 a, third lane). However, we failed to detect equally high levels of LMNB1 in cells with reduced LMNA/C, despite the fact that these cells were resistant to neomycin (neo). As shown in Fig. 5 b, increased expression of LMNB1 in cells with reduced LMNA/C (shLMNA/C:LMNB1) resulted in a markedly reduced rate of proliferation. This growth defect was more severe than the effect of LMNB1 overexpression in control cells. shLMNA/C:LMNB1 cells exhibited a flattened morphology (Fig. 5 c, bottom) and increased staining for SA-β-gal (Fig. 5 d). Furthermore, FACS analysis revealed that shLMNA/C:LMNB1 cells had a significantly reduced S-phase index and an increased G0/G1 peak, hallmarks of cells entering replicative senescence (Fig. 5 e). These results demonstrate that shLMNA/C:LMNB1 cells senesced. In Fig. 1 and Fig. S1, we showed that senescent cells had reduced LMNB1; the fact that we failed to detect high levels of LMNB1 in shLMNA/C:LMNB1 cells by Western analysis suggests that they down-regulated LMNB1 as a consequence of senescence (Fig. 5 a, fourth lane). shLMNA/C:LMNB1 cells also showed increased DNA damage compared with controls as measured by the numbers of 53BP-1 foci (Fig. 5 f). To investigate whether these DNA damage foci colocalize with telomeres, we costained the cells with TRF1 and 53BP-1 and scored for telomere dysfunction–induced DNA damage foci (TIFs). TIFs were previously identified in cells expressing TRF2ΔBΔM, which removes endogenous TRF2 from the telomeres and triggers a DNA damage response (Takai et al., 2003). TIFs were clearly identifiable in our positive control cells that expressed TRF2ΔBΔM (Fig. 5 g). Similarly, we observed many TIFs in shLMNA/C:LMNB1 cells compared with controls. Telomere-associated DNA damage foci often encompassed several telomeres (visualized by TRF1 staining), forming telomere aggregates (Fig. 5 g, bottom two rows). Quantification of TIFs revealed that the proportion of cells harboring one, two, or more than three TIFs was significantly increased in shLMNA/C:LMNB1 cells versus controls (Fig. 5 h). Conversely, the proportion of cells with no TIFs was reduced in shLMNA/C:LMNB1 cells. Because of the nature of telomeric chromatin, telomere-associated DNA damage foci persist and cannot be resolved by conventional DNA repair mechanisms under normal circumstances. Collectively, these results reveal that a reduction in LMNA/C enhances the consequences of increased LMNB1 expression, causing telomere dysfunction and impaired cell proliferation.
Discussion
Mutations in LMNA result in diseases affecting skeletal homeostasis, muscle, heart, vascular tissue, and premature aging, whereas duplication of the LMNB1 locus results in ADLD (Chen et al., 2003; De Sandre-Giovannoli et al., 2003; Eriksson et al., 2003; Csoka et al., 2004; Padiath and Fu, 2010). Increased levels of LMNB1 have also been observed in patients with AT (Barascu et al., 2012). Interestingly, both ADLD and AT affect myelination of the CNS, resulting in severe neurological defects.
Recent studies showed that LMNB1 loss is associated with senescence in fibroblasts (Shimi et al., 2011; Freund et al., 2012). Furthermore, alterations in LMNB1 levels affect cell proliferation and entry into replicative senescence (Shimi et al., 2011; Barascu et al., 2012). These findings raise the question whether the decline in LMNB1 during senescence occurs in other cell types or during aging in vivo.
Here, we show that both LMNB1 and LAP2 decrease during senescence of the two principal cell types of human skin: primary dermal fibroblasts and keratinocytes. These results are relevant in vivo, as LMNB1 decreases in keratinocytes during normal aging of human skin. Consequently, LMNB1 reduction may serve as a prognostic marker for skin aging and possibly other skin pathologies. These results also suggest that the nuclear lamina undergoes profound changes as cells enter replicative senescence.
Is loss of LMNB1 a cause or a consequence of cellular senescence? A recent study suggested that shRNA-mediated down-regulation of LMNB1 triggers senescence, in a manner that is reversible by p53 inactivation or growth under hypoxic conditions (Shimi et al., 2011). Here, we show that LMNB1 depletion, using constitutive or doxycyclin-inducible shRNA constructs, resulted in reduced proliferation in dermal fibroblasts (and their hTERT-immortalized counterparts) as well as in WI-38. This growth defect was not rescued by hTERT, p53 inactivation, or by growth under hypoxic conditions. In addition, we could not detect a significant increase in the number of senescent cells in LMNB1-depleted populations. However, we did observe an increase in SA-β-gal–positive cells when LMNB1-depleted cells were grown under sparse conditions. Based on these results, we conclude that LMNB1 depletion does not directly cause senescence unless it is accompanied by additional stress, such as sparse growth. Thus, LMNB1 loss is a hallmark and consequence but not a cause of senescence.
The importance of maintaining appropriate levels of LMNB1 is highlighted by the fact that in humans, an increase in LMNB1 expression, caused by duplication of the LMNB1 locus, is associated with ADLD (Padiath et al., 2006; Brussino et al., 2010). ADLD is characterized by widespread myelin loss in the CNS, reminiscent of chronic progressive multiple sclerosis. An increase in LMNB1 has also been observed in fibroblasts and lymphocytes from patients with AT, an autosomal-recessive disorder caused by mutations in the DNA damage response factor ATM (Barascu et al., 2012). Both ADLD and AT lead to demyelination of the CNS and acute neurological defects.
How do increased levels of LMNB1 lead to these detrimental cellular consequences, and why, as in ADLD, are these effects confined to the brain? We demonstrate that elevated LMNB1 levels lead to impaired proliferation and increased senescence in human fibroblasts. These phenotypes are rescued by expression of hTERT or by inactivation of the p53 tumor suppressor pathway. However, the phenotypic consequences of overexpressing physiologically relevant levels of LMNB1 in fibroblasts were subtle. This may not be unexpected, as ADLD patients do not show any overt skin problems. In contrast, neurological defects appear to be a consistent effect in humans and mice that have either increased or decreased LMNB1 levels, respectively (Padiath et al., 2006; Brussino et al., 2010; Coffinier et al., 2010, 2011).
Why is the CNS more susceptible to LMNB1 fluctuations? Is it possible that the low levels of LMNA/C in CNS cells (Zhang et al., 2011; Jung et al., 2012) renders them more susceptible to LMNB1 fluctuations? If so, would fibroblasts with reduced LMNA/C become more vulnerable to the effects of LMNB1 overexpression? Our results clearly demonstrate that this is indeed the case. Increased LMNB1 in cells with reduced LMNA/C resulted in a significantly greater proliferative defect than fibroblasts with normal levels of LMNA/C. This proliferation defect was accompanied by increased SA-β-gal staining, reduced S-phase index, an increase in the percentage of cells in the G0/G1 stage of the cell cycle, and greater DNA damage. In particular, shLMNA/C:LMNB1 fibroblasts exhibited increased telomeric DNA damage, suggesting that telomere dysfunction may be responsible for the proliferation defects and premature senescence observed in these cells. In conclusion, low levels of LMNA/C exacerbate the effects of increased LMNB1 in fibroblasts.
Our results may explain why LMNB1 alterations seem to mainly affect neuronal lineages and how low levels of LMNA/C may contribute to the pathology of ADLD. Additionally, other differences in the composition of the nuclear lamina or the cellular physiology of neurons versus fibroblasts may account for the tissue-specific nature of ADLD.
Impaired proliferation, increased senescence, and telomere-associated DNA damage that is rescued by hTERT have also been shown in cells expressing progerin, the truncated form of LMNA, which causes the premature aging syndrome HGPS (Kudlow et al., 2008; Benson et al., 2010). It remains unclear how increased LMNB1 expression or progerin independently results in growth arrest and cellular senescence. However, these observations raise important questions as to how perturbations to the nuclear lamina can interfere with human telomeres. This missing link may be crucial to understanding how mutations or stoichiometric changes in the levels of different lamins lead to such a wide variety of diseases and in turn may offer new routes to therapeutic intervention.
Materials and methods
Fibroblast and keratinocyte cell culture
All human material was obtained with informed donor consent and local ethical committee approval. Primary dermal fibroblasts were a gift from B. Reversade (Institute of Medical Biology, Singapore). Cells were grown under standard conditions (37°C; 5% CO2) in MEM (Invitrogen) supplemented with 50 U/ml penicillin and streptomycin (Invitrogen), 15% FCS (Invitrogen), 0.2 mM nonessential amino acids (Invitrogen), and 2 mM glutamine (Invitrogen). An original aliquot of the original LMNA E145K cell line was a gift from M. Eriksson (Karolinska Institute, Stockholm, Sweden). Primary keratinocyte cells were isolated from fresh skin samples using dispase and trypsin treatments to initiate a pure culture. Primary and N/TERT-1 keratinocyte cells were cultured in keratinocyte serum-free medium (Gibco) containing 25 µg/ml bovine pituitary extract, 0.1 mg/ml penicillin/streptomycin, 0.2 ng/ml epidermal growth factor, and 0.3 mM CaCl2 as previously described (Dickson et al., 2000; Invitrogen) or DermaLife Basal Medium + provided supplements (Lifeline Cell Technology). Sites of skin biopsies included scalp, eyelid, and skin above the upper lip.
Retro- and lentiviral constructs
pBABE-neo-hTERT, pBABE-puro-hTERT (Counter et al., 1998), pBABE-hygro, and pBABE-hygro-p53DD (Shaulian et al., 1992) were obtained from B. Weinberg (Massachusetts Institute of Technology, Cambridge, MA) via Addgene. pLPC-TRF2ΔBΔM (puro) was obtained from T. de Lange (Rockefeller University, New York, NY) via Addgene. LMNB1 was amplified from a human cDNA and cloned into pBABE-neo. Retroviruses were generated using standard procedures. Two different shRNAs against LMNB1 were obtained from Thermo Fisher Scientific. Target sequences for LMNB1 silencing were LMNB1A, 5′-GGCGAAGATGTGAAGGTTATA-3′; and LMNB1B, 5′-GGAGACACATCAGTCAGTTAT-3′. The target sequence for LMNA/C knockdown was 5′-TGCGCTTTTTGGTGACGCT-3′. Lentiviruses were generated according to the manufacturer’s protocol (Thermo Fisher Scientific). v5-LMNB1 was cloned into pTRIPZ (Thermo Fisher Scientific) by replacing the whole shRNA cassette with the full-length cDNA of human LMNB1. Stable cell lines were generated by selecting virally transduced cells for 7–12 d with their respective antibiotics: 1 µg/ml puro (Sigma-Aldrich), 0.05 mg/ml hygro (Sigma-Aldrich), and 0.1 mg/ml G418 (Life Technologies). pTRIPZ-based constructs (containing LMNB1 shRNAs or v5-tagged LMNB1) were induced with 1 µg/ml doxycyclin for 5 d before performing assays.
Immunofluorescence and image acquisition
Fibroblasts were grown on chambered coverglass slides (Lab-Tek; Thermo Fisher Scientific) for 2–3 d, fixed for 10 min in 4% paraformaldehyde, washed in PBS, permeabilized using PBS + 0.3% Triton X-100, and blocked in PBS + 5% FBS + 1% BSA. Cells were incubated with primary antibodies in blocking solution overnight at 4°C, washed in PBS, probed with secondary antibodies for 30–45 min at RT, and DAPI stained in PBS for 5 min. Images were acquired on an inverted microscope (Axiovert 200M; Carl Zeiss) using 10×, NA 0.3 Plan-Neofluar (Carl Zeiss), 40×, NA 0.60 Ph2 Corr long distance Plan-Neofluar, or 63×, NA 1.4 oil differential interference contrast Plan-Apochromat (Carl Zeiss) objectives and a camera (AxioCam MRm; Carl Zeiss). Images were processed and exported using AxioVision LE software 4.5 SP (2006; Carl Zeiss). Images were cropped, and figures were assembled using Photoshop (CS4; Adobe) and Illustrator (CS3; Adobe). Nuclear abnormalities (by Jol2 staining) and DNA damage (by 53BP and γ-H2A-X staining) were quantified by scoring 350–500 cells for each cell line and condition. Confocal imaging was performed on an upright confocal microscope (FV-1000; Olympus) using a 100× oil objective. TIFs were identified by colocalization of TRF1 with 53BP-1. Scoring criteria were zero, one, two, and more than three TIFs per nucleus.
Immunohistochemistry
Formalin-fixed, wax-embedded sections of skin biopsies were stained by immunoperoxidase after epitope heat retrieval at pH 6. After 1-h primary antibody incubation, sections were washed in tap water and incubated for 30 min with HRP-labeled polymer conjugated to goat anti–mouse immunoglobulins (Dako). Sections were developed with DAB substrate (Abcam) and counterstained with hematoxylin before dehydrating and mounting in DPX (CellPath). Photographs were taken with a microscope (Axio Imager Z1; Carl Zeiss) using a 40×, NA 0.60 Ph2 Corr long distance Plan-Neofluar objective.
Immunoblotting
Whole-cell lysates were generated using the Lysis-M kit solution (cOmplete; Roche). Lysates were quantified using the bicinchoninic acid protein assay kit (Microplate; Thermo Fisher Scientific), separated by SDS-PAGE, and transferred onto nitrocellulose membranes. Membranes were blocked for 1 h in Odyssey blocking buffer/PBS (1:1; LI-COR Biosciences) and hybridized with indicated antibodies overnight at 4°C. Membranes were washed in PBS + 0.1% Tween 20, and two-color detection was carried out using Odyssey infrared–labeled secondary antibodies. Membranes were scanned, and signals quantified using the scanner (Odyssey; LI-COR Biosciences).
Antibodies
Antibodies were used against the following proteins as indicated: LMNB1 (YenZym), LMNA/C and progerin (JoL2 and MAB3211; EMD Millipore), LMNB2 (LN43 and MAB3536; EMD Millipore), v5 tag (37–7500; Invitrogen), GAPDH (G9545; Sigma-Aldrich), β-tubulin (MRB 435P; Covance), β-actin (A5441; Sigma-Aldrich), 53BP-1 (NB100-304; Novus Biologicals), antiphospho–Histone H2A-X (Ser139 and 05–636; EMD Millipore), TRF2 (13579; Abcam), LAP2 (28541; Santa Cruz Biotechnology, Inc.), LAP2-α (5162; Abcam), and Ki-67 (MM-1; Novocastra). The p53 antibody was a gift from D. Lane (p53 Laboratory, A*STAR Biomedical Sciences Institutes, Singapore).
qRT-PCR
RNA was isolated using TRIZOL (Life Technologies) and further purified using the RNeasy kit (QIAGEN). DNase treatment was performed on a column, and total RNA was quantified by a spectrophotometer (ND-1000; NanoDrop Technologies). cDNA was generated using reverse transcription supermix for qRT-PCR (iScript; Bio-Rad Laboratories). For qRT-PCR, the TaqMan Fast Universal PCR master mix was used with TaqMan primers obtained from Applied Biosystems: LMNA/C (Hs00153462_m1), LMNB1 (Hs01059210_m1), and GAPDH (4326317E). qRT-PCR was performed on a real-time PCR system (7500 Fast; Applied Biosystems).
Growth curves and proliferation assays
Growth curves were performed using the xCELLigence System (Roche) in 96-well plates. Cell Index was monitored every hour throughout the experiment. For proliferation assays, cells were seeded in triplicates in 12-well culture plates and grown for 3–5 d under various conditions (hypoxia vs. normal O2 concentration), trypsinized, and counted using an automated cell counter (Scepter; EMD Millipore).
SA-β-gal staining
Cells were fixed in 2% formaldehyde and 0.2% glutaraldehyde for 5 min at RT, washed twice in PBS, and incubated for 6 h in 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside as described previously (Dimri et al., 1995).
miR-23a qRT-PCR detection and luciferase reporter assays
Quantification of miR-23a levels was performed using the TaqMan miRNA reverse transcription kit. The expression level of miR-23a was measured using gene-specific predesigned TaqMan primers. U6 was used as a normalization control. The 3′UTR from human LMNB1 was cloned into psiCHECK2 vector, and mutations were introduced using PCR-based mutagenesis. Mutations were verified by sequencing. The miR-23a precursor plus an ∼300-bp flanking region was amplified from human genomic DNA and inserted into miRVecBlast using BamHI and EcoRI restriction sites. miRVecblast-miR-882 was a gift from D. Zhang (Institute of Medical Biology, Singapore). For transfection, 50 ng psiCHECK2 plasmid DNA and 500 ng miRVecBlast plasmid DNA were cotransfected into each well of a 24-well plate using transfection reagent (Lipofectamine 2000; Invitrogen) and 35 µl Opti-MEM (Invitrogen). Duplicates were made, and cells were harvested 40 h after transfection. Samples were lysed and harvested to determine luciferase activity. Sample signals were detected using a detection kit (Dual Luciferase; Promega). To determine the effects of miR-23a or miR-882 on LMNB1 protein and mRNA levels, 500 ng miRNA (in miRVecBlast) was transfected into dermal fibroblasts using Lipofectamine 2000. mRNA and protein lysates were harvested 30 h after transfection. Experiments were run in biological triplicates. P-values were determined using the Student’s t test.
Online supplemental material
Fig. S1 provides evidence of LMNB1 loss in progeria patient-derived fibroblasts, normal senescent human fibroblasts, and upon induction of telomere-specific DNA damage. Fig. S2 shows that the lamina-associated polypeptide (LAP2) α isoform also declines in senescent fibroblasts. Fig. S3 demonstrates that in senescent cells, loss of LMNB1 mRNA is accompanied by increased levels of miR-23a. Fig. S4 shows that in contrast to LMNB1-depleted cells, LMNB2 loss does not result in any overt proliferation phenotype. Fig. S5 provides information regarding our lentiviral transduction efficiency, SA-β-gal staining, and quantification of SA-β-gal–positive cells in LMNB1-depleted or normal human fibroblasts grown at different seeding densities. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201206121/DC1.
Supplementary Material
Acknowledgments
We are grateful to SeongSoo Lim, Wei Yong Chua, and Valere Chacheux-Rataboul (Genome Institute of Singapore) for karyotypic analysis of LMNB1-deficient cells. We thank Brian Burke, Philipp Kaldis, and Sebastien Tessier for insightful discussions and Maria Eriksson for providing us with LMNA E145K cells. Graham Wright and the Institute of Medical Biology microscopy unit are thanked for help with image acquisition. The authors would like to thank the anonymous reviewers for their thorough review and insightful comments on this work.
This work was supported by the Singapore Biomedical Research Council of the Singapore Agency for Science, Technology and Research (A*STAR).
Footnotes
Abbreviations used in this paper:
- ADLD
- adult-onset autosomal dominant leukodystrophy
- AT
- ataxia telangiectasia
- CNS
- central nervous system
- HGPS
- Hutchinson-Gilford progeria syndrome
- hTERT
- human TERT
- PD
- population doubling
- qRT-PCR
- quantitative RT-PCR
- SA-β-gal
- senescence-associated β-galactosidase
- TERT
- telomerase reverse transcription
- TIF
- telomere dysfunction–induced DNA damage focus
References
- Barascu A., Le Chalony C., Pennarun G., Genet D., Imam N., Lopez B., Bertrand P. 2012. Oxidative stress induces an ATM-independent senescence pathway through p38 MAPK-mediated lamin B1 accumulation. EMBO J. 31:1080–1094 10.1038/emboj.2011.492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson E.K., Lee S.W., Aaronson S.A. 2010. Role of progerin-induced telomere dysfunction in HGPS premature cellular senescence. J. Cell Sci. 123:2605–2612 10.1242/jcs.067306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridger J.M., Kill I.R. 2004. Aging of Hutchinson-Gilford progeria syndrome fibroblasts is characterised by hyperproliferation and increased apoptosis. Exp. Gerontol. 39:717–724 10.1016/j.exger.2004.02.002 [DOI] [PubMed] [Google Scholar]
- Brussino A., Vaula G., Cagnoli C., Panza E., Seri M., Di Gregorio E., Scappaticci S., Camanini S., Daniele D., Bradac G.B., et al. 2010. A family with autosomal dominant leukodystrophy linked to 5q23.2-q23.3 without lamin B1 mutations. Eur. J. Neurol. 17:541–549 10.1111/j.1468-1331.2009.02844.x [DOI] [PubMed] [Google Scholar]
- Burke B., Stewart C.L. 2006. The laminopathies: the functional architecture of the nucleus and its contribution to disease. Annu. Rev. Genomics Hum. Genet. 7:369–405 10.1146/annurev.genom.7.080505.115732 [DOI] [PubMed] [Google Scholar]
- Campisi J., d’Adda di Fagagna F. 2007. Cellular senescence: when bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 8:729–740 10.1038/nrm2233 [DOI] [PubMed] [Google Scholar]
- Chen L., Lee L., Kudlow B.A., Dos Santos H.G., Sletvold O., Shafeghati Y., Botha E.G., Garg A., Hanson N.B., Martin G.M., et al. 2003. LMNA mutations in atypical Werner’s syndrome. Lancet. 362:440–445 10.1016/S0140-6736(03)14069-X [DOI] [PubMed] [Google Scholar]
- Coffinier C., Chang S.Y., Nobumori C., Tu Y., Farber E.A., Toth J.I., Fong L.G., Young S.G. 2010. Abnormal development of the cerebral cortex and cerebellum in the setting of lamin B2 deficiency. Proc. Natl. Acad. Sci. USA. 107:5076–5081 10.1073/pnas.0908790107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffinier C., Jung H.J., Nobumori C., Chang S., Tu Y., Barnes R.H., II, Yoshinaga Y., de Jong P.J., Vergnes L., Reue K., et al. 2011. Deficiencies in lamin B1 and lamin B2 cause neurodevelopmental defects and distinct nuclear shape abnormalities in neurons. Mol. Biol. Cell. 22:4683–4693 10.1091/mbc.E11-06-0504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counter C.M., Hahn W.C., Wei W., Caddle S.D., Beijersbergen R.L., Lansdorp P.M., Sedivy J.M., Weinberg R.A. 1998. Dissociation among in vitro telomerase activity, telomere maintenance, and cellular immortalization. Proc. Natl. Acad. Sci. USA. 95:14723–14728 10.1073/pnas.95.25.14723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csoka A.B., Cao H., Sammak P.J., Constantinescu D., Schatten G.P., Hegele R.A. 2004. Novel lamin A/C gene (LMNA) mutations in atypical progeroid syndromes. J. Med. Genet. 41:304–308 10.1136/jmg.2003.015651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Adda di Fagagna F., Reaper P.M., Clay-Farrace L., Fiegler H., Carr P., Von Zglinicki T., Saretzki G., Carter N.P., Jackson S.P. 2003. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 426:194–198 10.1038/nature02118 [DOI] [PubMed] [Google Scholar]
- Decker M.L., Chavez E., Vulto I., Lansdorp P.M. 2009. Telomere length in Hutchinson-Gilford progeria syndrome. Mech. Ageing Dev. 130:377–383 10.1016/j.mad.2009.03.001 [DOI] [PubMed] [Google Scholar]
- de Lange T. 2005. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 19:2100–2110 10.1101/gad.1346005 [DOI] [PubMed] [Google Scholar]
- De Sandre-Giovannoli A., Bernard R., Cau P., Navarro C., Amiel J., Boccaccio I., Lyonnet S., Stewart C.L., Munnich A., Le Merrer M., Lévy N. 2003. Lamin a truncation in Hutchinson-Gilford progeria. Science. 300:2055 10.1126/science.1084125 [DOI] [PubMed] [Google Scholar]
- Dickson M.A., Hahn W.C., Ino Y., Ronfard V., Wu J.Y., Weinberg R.A., Louis D.N., Li F.P., Rheinwald J.G. 2000. Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol. Cell. Biol. 20:1436–1447 10.1128/MCB.20.4.1436-1447.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimri G.P., Lee X., Basile G., Acosta M., Scott G., Roskelley C., Medrano E.E., Linskens M., Rubelj I., Pereira-Smith O., et al. 1995. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. USA. 92:9363–9367 10.1073/pnas.92.20.9363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner D., Vlcek S., Foeger N., Gajewski A., Makolm C., Gotzmann J., Hutchison C.J., Foisner R. 2006. Lamina-associated polypeptide 2α regulates cell cycle progression and differentiation via the retinoblastoma–E2F pathway. J. Cell Biol. 173:83–93 10.1083/jcb.200511149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson M., Brown W.T., Gordon L.B., Glynn M.W., Singer J., Scott L., Erdos M.R., Robbins C.M., Moses T.Y., Berglund P., et al. 2003. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 423:293–298 10.1038/nature01629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund A., Laberge R.M., Demaria M., Campisi J. 2012. Lamin B1 loss is a senescence-associated biomarker. Mol. Biol. Cell. 23:2066–2075 10.1091/mbc.E11-10-0884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelen L., Pagie L., Brasset E., Meuleman W., Faza M.B., Talhout W., Eussen B.H., de Klein A., Wessels L., de Laat W., van Steensel B. 2008. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 453:948–951 10.1038/nature06947 [DOI] [PubMed] [Google Scholar]
- Hahn W.C., Dessain S.K., Brooks M.W., King J.E., Elenbaas B., Sabatini D.M., DeCaprio J.A., Weinberg R.A. 2002. Enumeration of the simian virus 40 early region elements necessary for human cell transformation. Mol. Cell. Biol. 22:2111–2123 10.1128/MCB.22.7.2111-2123.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harborth J., Elbashir S.M., Bechert K., Tuschl T., Weber K. 2001. Identification of essential genes in cultured mammalian cells using small interfering RNAs. J. Cell Sci. 114:4557–4565 [DOI] [PubMed] [Google Scholar]
- Huntzinger E., Izaurralde E. 2011. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet. 12:99–110 10.1038/nrg2936 [DOI] [PubMed] [Google Scholar]
- Jung H.J., Coffinier C., Choe Y., Beigneux A.P., Davies B.S., Yang S.H., Barnes R.H., II, Hong J., Sun T., Pleasure S.J., et al. 2012. Regulation of prelamin A but not lamin C by miR-9, a brain-specific microRNA. Proc. Natl. Acad. Sci. USA. 109:E423–E431 10.1073/pnas.1111780109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlseder J., Broccoli D., Dai Y., Hardy S., de Lange T. 1999. p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science. 283:1321–1325 10.1126/science.283.5406.1321 [DOI] [PubMed] [Google Scholar]
- Kim Y., Sharov A.A., McDole K., Cheng M., Hao H., Fan C.M., Gaiano N., Ko M.S., Zheng Y. 2011. Mouse B-type lamins are required for proper organogenesis but not by embryonic stem cells. Science. 334:1706–1710 10.1126/science.1211222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudlow B.A., Stanfel M.N., Burtner C.R., Johnston E.D., Kennedy B.K. 2008. Suppression of proliferative defects associated with processing-defective lamin A mutants by hTERT or inactivation of p53. Mol. Biol. Cell. 19:5238–5248 10.1091/mbc.E08-05-0492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.T., Fu Y.H. 2009. miR-23 regulation of lamin B1 is crucial for oligodendrocyte development and myelination. Dis Model Mech. 2:178–188 10.1242/dmm.001065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G.H., Barkho B.Z., Ruiz S., Diep D., Qu J., Yang S.L., Panopoulos A.D., Suzuki K., Kurian L., Walsh C., et al. 2011. Recapitulation of premature ageing with iPSCs from Hutchinson-Gilford progeria syndrome. Nature. 472:221–225 10.1038/nature09879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhas A.N., Lee C.F., Vaux D.J. 2009. Lamin B1 controls oxidative stress responses via Oct-1. J. Cell Biol. 184:45–55 10.1083/jcb.200804155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir R.D., Montag-Lowy M., Goldman R.D. 1994. Dynamic properties of nuclear lamins: lamin B is associated with sites of DNA replication. J. Cell Biol. 125:1201–1212 10.1083/jcb.125.6.1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naetar N., Korbei B., Kozlov S., Kerenyi M.A., Dorner D., Kral R., Gotic I., Fuchs P., Cohen T.V., Bittner R., et al. 2008. Loss of nucleoplasmic LAP2alpha-lamin A complexes causes erythroid and epidermal progenitor hyperproliferation. Nat. Cell Biol. 10:1341–1348 10.1038/ncb1793 [DOI] [PubMed] [Google Scholar]
- Padiath Q.S., Fu Y.H. 2010. Autosomal dominant leukodystrophy caused by lamin B1 duplications a clinical and molecular case study of altered nuclear function and disease. Methods Cell Biol. 98:337–357 10.1016/S0091-679X(10)98014-X [DOI] [PubMed] [Google Scholar]
- Padiath Q.S., Saigoh K., Schiffmann R., Asahara H., Yamada T., Koeppen A., Hogan K., Ptácek L.J., Fu Y.H. 2006. Lamin B1 duplications cause autosomal dominant leukodystrophy. Nat. Genet. 38:1114–1123 10.1038/ng1872 [DOI] [PubMed] [Google Scholar]
- Pekovic V., Harborth J., Broers J.L., Ramaekers F.C., van Engelen B., Lammens M., von Zglinicki T., Foisner R., Hutchison C., Markiewicz E. 2007. Nucleoplasmic LAP2α–lamin A complexes are required to maintain a proliferative state in human fibroblasts. J. Cell Biol. 176:163–172 10.1083/jcb.200606139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röber R.A., Weber K., Osborn M. 1989. Differential timing of nuclear lamin A/C expression in the various organs of the mouse embryo and the young animal: a developmental study. Development. 105:365–378 [DOI] [PubMed] [Google Scholar]
- Scaffidi P., Misteli T. 2005. Reversal of the cellular phenotype in the premature aging disease Hutchinson-Gilford progeria syndrome. Nat. Med. 11:440–445 10.1038/nm1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaulian E., Zauberman A., Ginsberg D., Oren M. 1992. Identification of a minimal transforming domain of p53: negative dominance through abrogation of sequence-specific DNA binding. Mol. Cell. Biol. 12:5581–5592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimi T., Butin-Israeli V., Adam S.A., Hamanaka R.B., Goldman A.E., Lucas C.A., Shumaker D.K., Kosak S.T., Chandel N.S., Goldman R.D. 2011. The role of nuclear lamin B1 in cell proliferation and senescence. Genes Dev. 25:2579–2593 10.1101/gad.179515.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart C., Burke B. 1987. Teratocarcinoma stem cells and early mouse embryos contain only a single major lamin polypeptide closely resembling lamin B. Cell. 51:383–392 10.1016/0092-8674(87)90634-9 [DOI] [PubMed] [Google Scholar]
- Taimen P., Pfleghaar K., Shimi T., Möller D., Ben-Harush K., Erdos M.R., Adam S.A., Herrmann H., Medalia O., Collins F.S., et al. 2009. A progeria mutation reveals functions for lamin A in nuclear assembly, architecture, and chromosome organization. Proc. Natl. Acad. Sci. USA. 106:20788–20793 10.1073/pnas.0911895106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai H., Smogorzewska A., de Lange T. 2003. DNA damage foci at dysfunctional telomeres. Curr. Biol. 13:1549–1556 10.1016/S0960-9822(03)00542-6 [DOI] [PubMed] [Google Scholar]
- Tang C.W., Maya-Mendoza A., Martin C., Zeng K., Chen S., Feret D., Wilson S.A., Jackson D.A. 2008. The integrity of a lamin-B1-dependent nucleoskeleton is a fundamental determinant of RNA synthesis in human cells. J. Cell Sci. 121:1014–1024 10.1242/jcs.020982 [DOI] [PubMed] [Google Scholar]
- Tsai M.Y., Wang S., Heidinger J.M., Shumaker D.K., Adam S.A., Goldman R.D., Zheng Y. 2006. A mitotic lamin B matrix induced by RanGTP required for spindle assembly. Science. 311:1887–1893 10.1126/science.1122771 [DOI] [PubMed] [Google Scholar]
- Vergnes L., Péterfy M., Bergo M.O., Young S.G., Reue K. 2004. Lamin B1 is required for mouse development and nuclear integrity. Proc. Natl. Acad. Sci. USA. 101:10428–10433 10.1073/pnas.0401424101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worman H.J., Ostlund C., Wang Y. 2010. Diseases of the nuclear envelope. Cold Spring Harb. Perspect. Biol. 2:a000760 10.1101/cshperspect.a000760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Lian Q., Zhu G., Zhou F., Sui L., Tan C., Mutalif R.A., Navasankari R., Zhang Y., Tse H.F., et al. 2011. A human iPSC model of Hutchinson Gilford Progeria reveals vascular smooth muscle and mesenchymal stem cell defects. Cell Stem Cell. 8:31–45 10.1016/j.stem.2010.12.002 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.