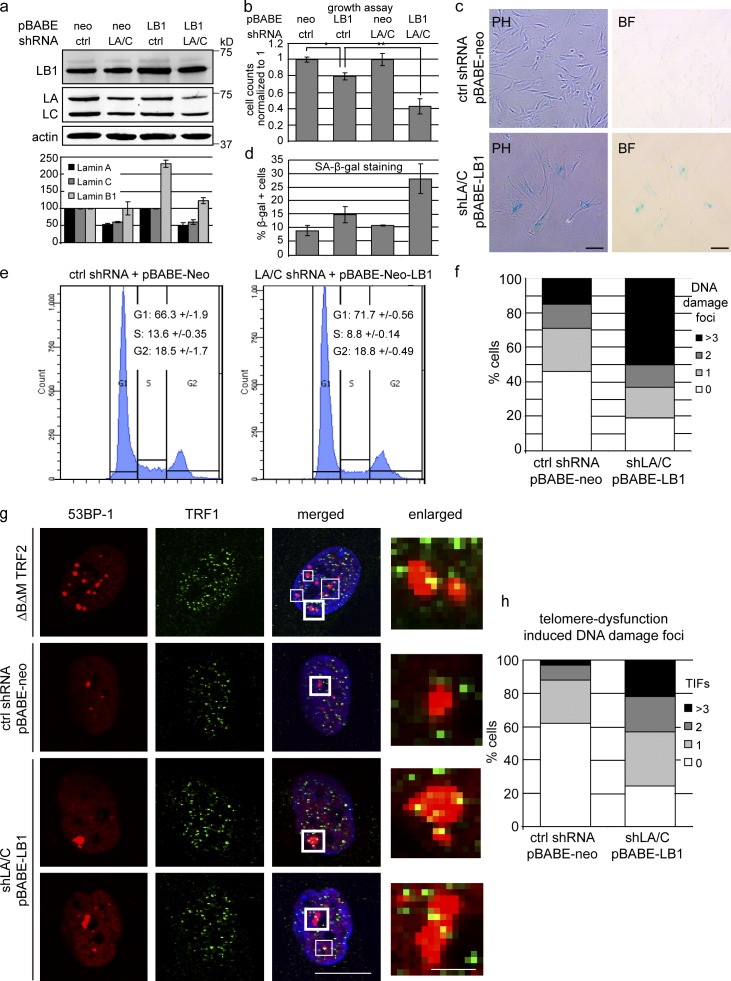
Figure 5.
Reduced levels of LMNA/C exacerbate the effects of LMMB1 overexpression. (a, top) Western blot showing LMNB1 (LB1) and LMNA/C (LA/C) levels in cells expressing pBABE-neo control or pBABE-neo-LMNB1 in cells previously transduced with control (ctrl) or LMNA/C shRNA. Antibodies are LMNB1, LMNA/C, and actin. (bottom) Quantification of LMNB1 and LMNA/C levels normalized to actin (n = 3). Data are from three independent experiments. (b) Proliferation assay of cells expressing pBABE-neo or pBABE-neo-LMNB1 in cells expressing control shRNA (first two bars) or in cells expressing shRNA against LMNA/C (last two bars). *, P < 0.05; **, P < 0.01; n = 4. (c) SA-β-gal staining of control versus LMNB1 in shLMNA/C cells (PH, phase contrast; BF, bright field). Bars, 50 µm. (d) Quantification of SA-β-gal–positive cells in cells described in b. (n = 4). (e) Cell cycle analysis of control versus LMNB1 in shLMNA/C cells. Percentages of cells in different cell cycle stages are indicated (n = 4). (f) Quantification of DNA damage foci by 53BP1 staining in control versus LMNB1 in shLMNA/C cells. At least 400 cells were counted for each condition. (g) Confocal immunofluorescence microscopy showing telomere dysfunction–induced DNA damage foci (TIFs). Cells were stained with 53BP-1 (red) and TRF1 (green). Shown are cells expressing TRF2ΔBΔM (positive control) or vector control (scrambled shRNA + pBABE-neo) and cells with reduced levels of LMNA/C overexpressing LMNB1 (bottom two rows). Merged images also show DAPI staining. Bar, 20 µm. White boxes highlight TIFs. Thick framed white boxes are enlarged on the right. Bar, 2.5 µm. (h) Quantification of TIFs from control versus shLMNA/C:LMNB1 cells. Percentages of cells showing zero, one, two or more than three TIFs are shown. 100 cells were counted for each condition. Error bars represent ± SD.