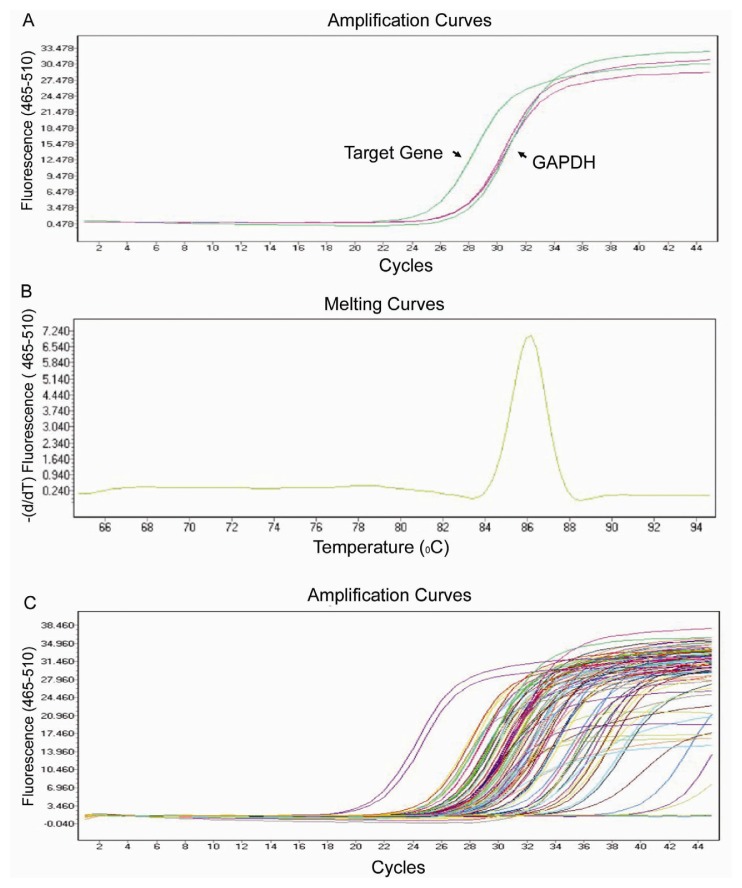
Figure 1.
Design and testing of the real-time PCR array for human genes encoding epigenetic chromatin modification enzymes. (A) Amplification of a target gene and GAPDH in the real-time PCR array. Reactions were run on a Light cycler 480 (Roche, Basel, Switzerland) using universal thermal cycling parameters (95 °C for 5 min, 45 cycles of 10 s at 95 °C, 20 s at 60 °C and 15 s at 72 °C); (B) Melting curve analysis of the PCR product of a single target gene. Melting curves were generated using the parameters 10 s at 95 °C, 60 s at 60 °C, followed by continued melting; (C) Amplification of all of the genes in the PCR array.