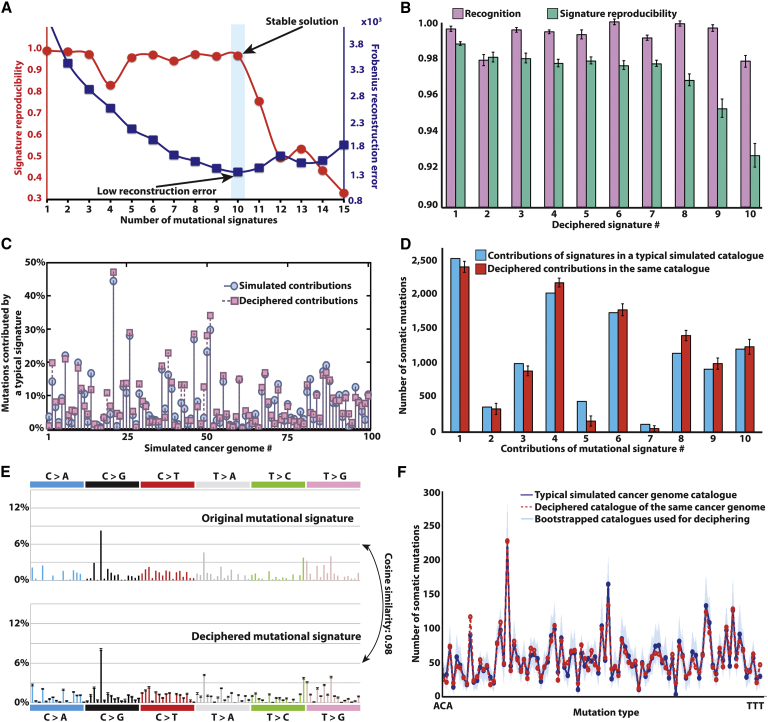
Figure 2.
Deciphering Signatures of Mutational Processes from a Set of Simulated Mutational Catalogs from 100 Cancer Genomes
(A) Identifying the number of processes operative in a set of 100 simulated cancer genomes based on reproducibility of their signatures and low error for reconstructing the original catalogs.
(B) Comparison between the ten deciphered signatures and the ten signatures used to simulate the catalogs. Signature recognition, measured using cosine similarity, and signature reproducibility, measured using average silhouette width, is given for each mutational signature. The error bars represent the SD of the corresponding characteristics for the extracted signature(s).
(C) Comparison between deciphered and simulated contributions of one of the ten mutational processes in all cancer genomes.
(D) Comparison between deciphered and simulated contributions of all signatures in a typical cancer genome. The error bars represent the SD of the corresponding characteristics for the extracted signature(s).
(E) Comparison between the profiles of typical deciphered and simulated signature. The error bars represent the SD of the corresponding characteristics for the extracted signature(s).
(F) Comparison between the mutational catalogs of a typical deciphered (red line) and simulated (dark blue line) cancer genome. The separately bootstrapped per iteration mutational catalogs (Experimental Procedures), which are used to decipher the mutational signatures and their contributions, are shown in light blue.