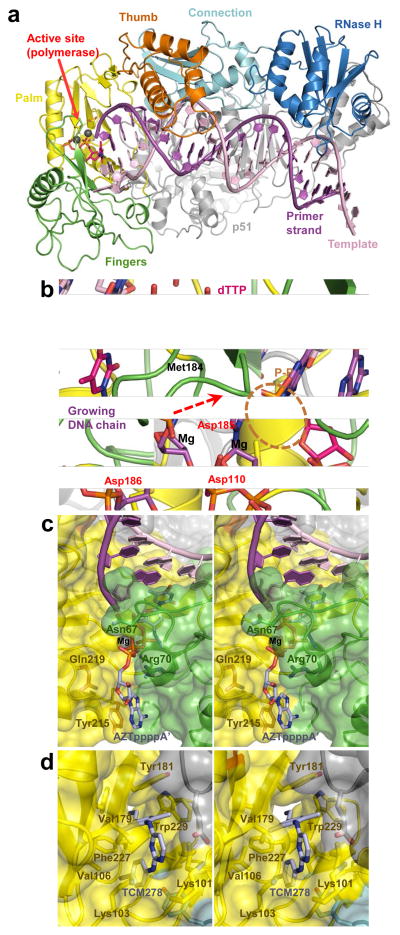
Figure 4.
Structural analyses of HIV-1 RT function and its inhibition by small molecules. (a) Overview of the HIV-1 RT-template-primer complex (pdb code 1RTD). The protein and DNA chains are shown as cartoons. The subdomains of the active RT subunit are indicated and colour-coded; the inactive (p51) subunit is shown in gray. The structure contains a bound molecule of dTTP in the active site (pink). Grey spheres are Mg atoms. (b) Close-up of the polymerase active site (pdb code 1RTD) and DNA polymerization. The 3′-hydroxyl, absent in the original structure 48, is added for illustration purposes. The direction of nucleophilic attack is indicated by a dashed arrow. The catalytic residues, Met184 and the leaving pyrophosphate group (P–P) are shown as sticks and indicated. RT chain colours are conserved from panel a. (c) Stereo view of ATP-binding pocket in AZT-resistant HIV-1 RT (pdb code 3KLE). The excision product (AZTpppp A′) is shown as sticks with carbon atoms in light blue. Protein chains are shown as cartoons with semitransparent surfaces (colouring as in panel a); residues implicated in AZT resistance are indicated. (d) TCM278 bound to HIV-1 RT (pdb code 2ZD1). RTresidues forming the NNRTI-binding pocket are indicated.