Figure 5.
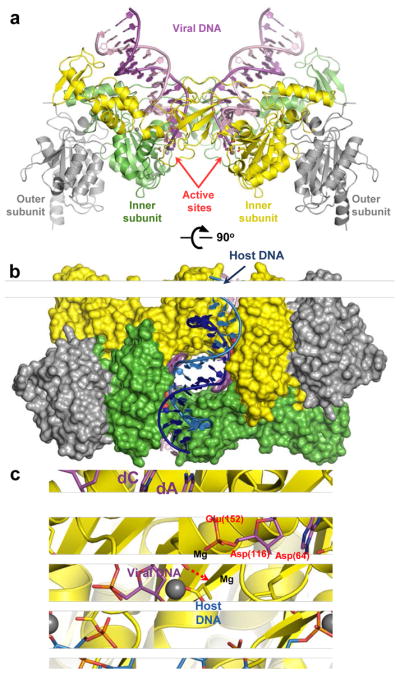
Retroviral intasome structures and mechanism of IN catalysis. (a) Overview of the PFV intasome structure (pdb code 3OY9). The active (inner) IN chains are shown as green and yellow cartoons; catalytically inactive (outer) chains are gray. The transferred and non-transferred viral DNA strands are shown in dark and light magenta, respectively. Active site carboxylates are shown as sticks and divalent metal ions as gray spheres. (b) The PFV intasome in complex with a host DNA mimic (light and dark blue; pdb code 3OS2). IN chains are shown in space-fill mode conserving colours from panel a. (c) DNA strand transfer. The model is based on structures of the Mn2+-bound intasome and target capture complex (see 84 for details). IN is shown as cartoons with D, D-35-E active site residues as sticks. DNA is shown as sticks; the invariant viral dA and dC nucleotides are indicated. Colours are conserved from panel a. Residue numbering corresponds to the HIV-1 IN sequence. Direction of nucleophilic attack is indicated by a red dashed arrow.
