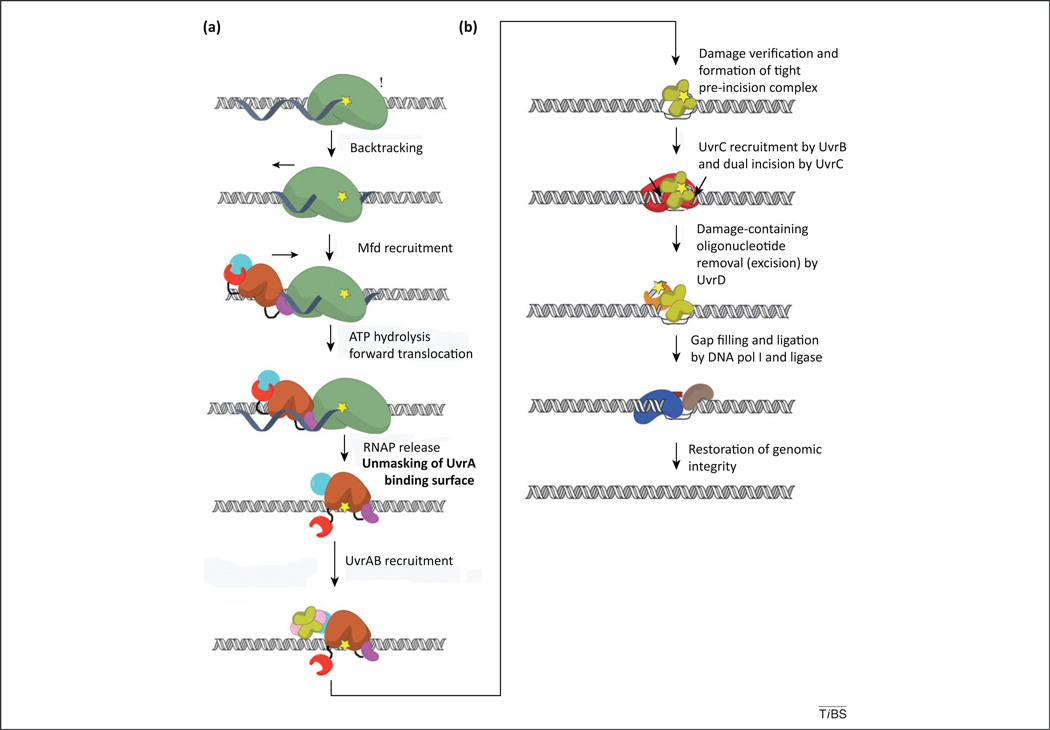
Figure 1.
Schematic representation of bacterial transcription-coupled DNA repair (TCR). Distinct modalities exist for dealing with DNA damage encountered by the transcription machinery, which can (i) release, (ii) reposition, or (iii) destroy the stalled RNA polymerase (RNAP) in order to expose the lesion to nucleotide excision repair (NER) proteins. Bacterial, Mfd-dependent TCR is an example of (i) [44], whereas eukaryotic TCR might occur by either (ii) or (iii) [8]. Mfd-dependent repair occurs through the following sequence of events: (a) elongating RNAP (green) stalls at DNA lesions in the template strand (yellow star) and might backtrack; this recruits Mfd [colored by module in cyan (D1–D3), magenta (RNAP-interacting domain; RID), brown (D5–D6), and red (D7)], which promotes forward translocation of RNAP through ATP hydrolysis by the translocase module. Annealing of the upstream edge of the transcription bubble eventually results in its collapse and ternary elongation complex (TEC) dissociation. Next, the UvrAB complex (the UvrB dimer is lime and UvrA is pink) is recruited by virtue of the unmasking of the UvrA-binding surface in D2 (cyan) by motion of D7. (b) The TCR pathway continues with formation of an UvrB–DNA preincision complex, recruitment of UvrC (red) by UvrB and dual incision of the DNA. The damage-containing oligonucleotide is then excised and removed via UvrD (orange), after which the gap is filled via the action of DNA polI (blue) and ligase (coffee). Global NER differs in the initial localization of the damage by the UvrAB complex, which does not depend on RNAP (as depicted in a), while subsequent to formation of the UvrB–DNA preincision complex (depicted in b), both global NER and TCR are thought to occur similarly. For simplicity, the UvrB preincision complex is depicted with two UvrB copies; however, given the competitive nature of UvrB/Mfd binding to UvrA [16,44], that remains a matter of debate.