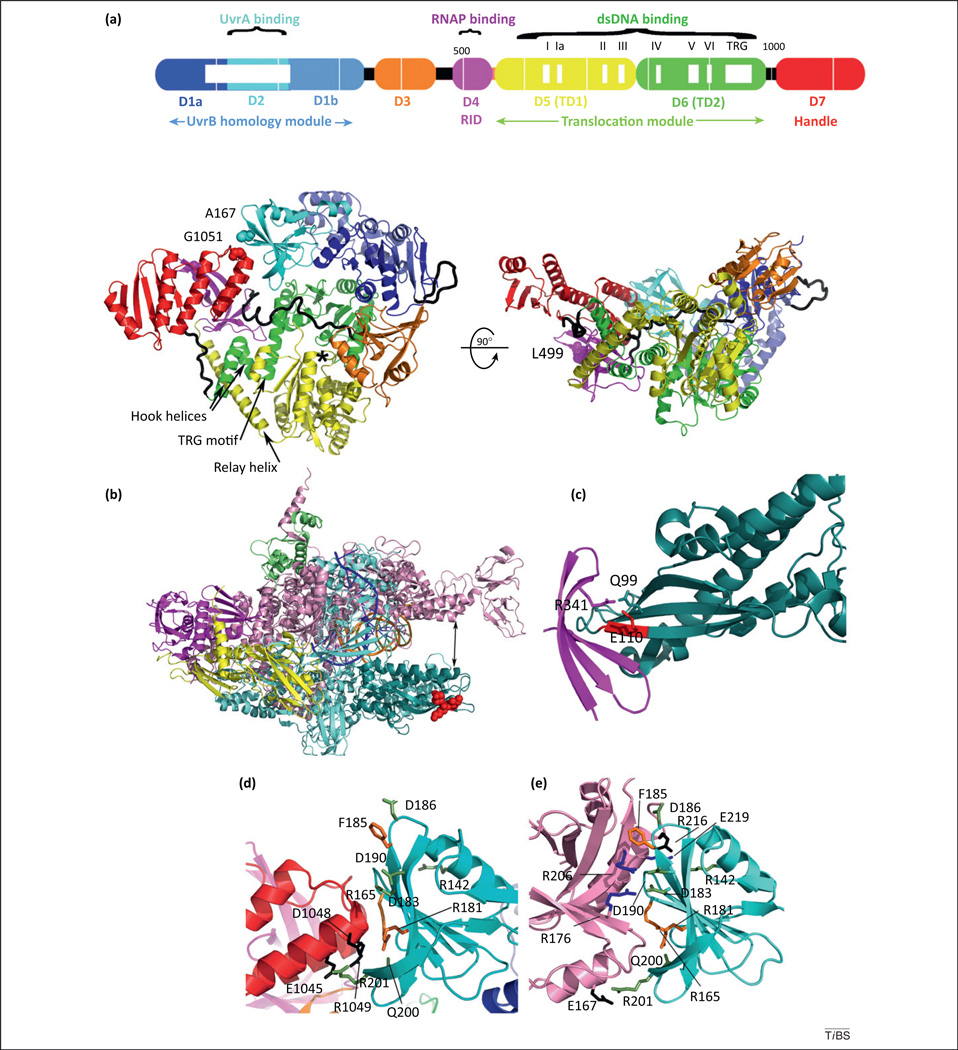
Figure 2.
Mfd structure and selected structural interfaces involved in transcription-coupled DNA repair (TCR). (a) Domain organization and crystal structure of Escherichia coli Mfd (PDB ID 2EYQ) seen from the top (left) and sideways (right). The molecule adopts a modular architecture with functional modules connected by flexible loops and hinges [12], shown as black tubes. Within the RNA polymerase (RNAP)-interacting domain (RID), substitution of L499 (spheres) compromises TCR in vivo and in vitro due to a defect of this variant in binding to RNAP [12]. Also shown in spheres are A167 and G1051, which are substituted for cysteine in order to create an intramolecular disulfide tethering D2 to D7 and resulting in masking of the UvrA binding surface [seen in (e)] located at the interface between D2 and D7 [16]. An asterisk indicates the ATP binding site and mobile structural elements presumed to change conformation during the mechanochemical cycle are labeled. Adapted from [12]. (b) Crystal structure of Thermus thermophilus ternary elongation complex assembled on a synthetic nucleic acid scaffold (PDB ID 2O5I) color coded by subunit as follows: β (cyan), β′ (pink), α (magenta and yellow), ω (olive), nascent RNA (blue), template dsDNA (orange). The crab-claw-shaped enzyme is positioned such that the downstream DNA is shown going into the page. Shown in teal is the β1 fragment that associates with the RID domain of Mfd [19]. The IKE sequence motif within the β1 fragment (initially identified in E. coli as I117 K118 E119) [29] and corresponding to Thermus aquaticus and T. thermophilus I108 K109 E110 [28]) is important for the Mfd–RNAP interaction and is shown as red spheres. (c) Close-up view of the hybrid T. thermophilus Mfd–RID and T. aquaticus RNAP-β1 fragment (PDB ID 3MLQ) [28]. The interaction is bipartite: the core interaction comprises evolutionarily conserved residues located on the edge β-strands and forming an intermolecular β-sheet; a second, phylum-specific peripheral interaction is mediated by R341 (corresponding to E. coli L499), and contacts the conserved IKE motif (red) and phylum-specific Q99. (d) Close-up view of the Mfd D2–D7 interaction inhibitory for UvrA binding. Conserved D7 residues are shown in black; D2 residues that support the interaction are shown in lime. Substitutions of orange residues are functionally important for TCR in vivo and in vitro [54]. Reproduced from [16]. (e) Close-up of the transcription–repair coupling factor (TRCF)–UvrA interface. Residues in UvrA that bind UvrB are shown in blue [59]. Other residues are colored as in (d). Reproduced from [16].