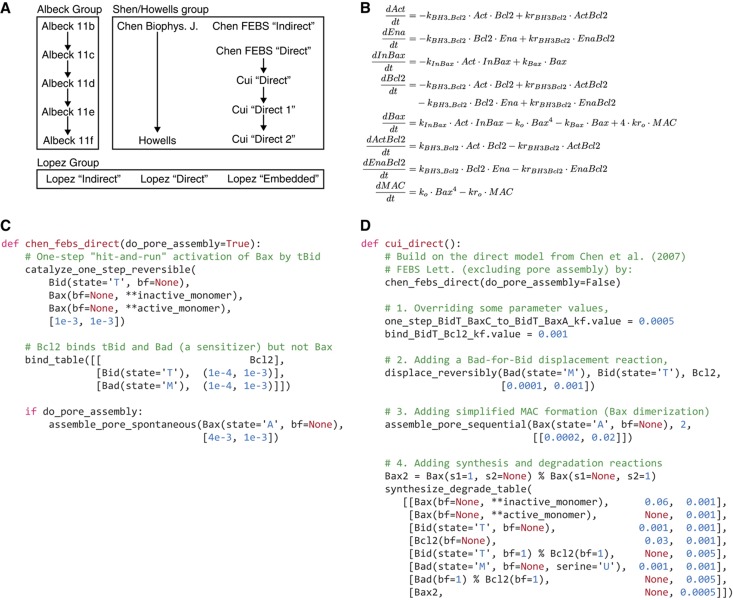
Refactoring of published models into PySB. (
A) Relationships between
the models examined in this paper. The ‘Albeck Group’
incorporates a series of incrementally expanded models shown in Figure 11 of
Albeck et al (2008b); the
‘Shen/Howells Group’ incorporates models from three papers from
the research group of Shen and colleagues (
Chen et
al, 2007a,
Chen et al,
2007b;
Cui et al, 2008)
and a derivative model from
Howells et al
(2010); the ‘Lopez group’ includes three expanded
models introduced in this paper. The arrows indicate that one model has been
derived or extended from a prior model and point in the direction Base Model
→ Derivative Model. (
B) The ‘Direct’ model from
Chen et al (2007b) in its original
ODE-based representation. (
C) Conversion of the Chen
‘Direct’ model to a PySB module. The execution of the
chen_febs_direct
function results in rules that exactly reproduce the ODEs
shown above (the molecule type
Bad
in the PySB function corresponds to the
generic enabler species
Ena in the original equations;
Bid
corresponds to the generic activator
Act). The macro
catalyze_one_step_reversible
implements the two-reaction scheme
E+S→E+P,
P→S;
assemble_pore_spontaneous
implements the order-4 reaction 4 ×
subunit
 pore
pore. The
bind_table
macro is illustrated in
Figure
2C. (
D) Model extension in PySB. Module Reuse (
Figure 3B) was used to implement the
‘Direct’ model from
Cui et al
(2008) as an extension of the prior ‘Direct’ model
from
Chen et al (2007b) shown in
Figure 4C. Invocation of the PySB function
chen_febs_direct
incorporates the elements of the original
Chen et al (2007b) model; subsequent
statements specify the modifications and additions required to yield the
derived model from
Cui et al (2008).
 pore. The
pore. The