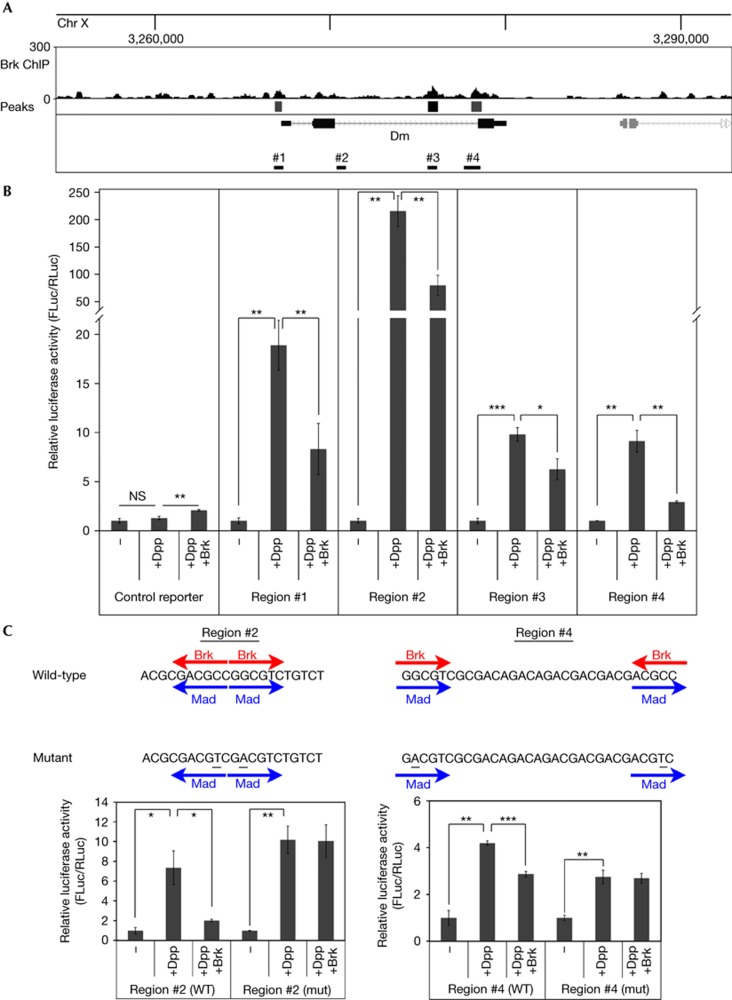
Figure 2.
The Myc genomic region contains elements responsive to Mad/Medea and Brk. (A) Brk binding in the Myc (dm) genomic region by ChIP-Seq. Brk binding peaks identified by the PeakSeq algorithm (‘Peaks’), indicated as boxes below the ChIP-seq trace. One peak (black box, also indicted as region #3 below the gene model) passes the threshold P<10−10. Two more peaks (grey boxes, regions #1 and #4) pass the less stringent default PeakSeq setting of P<0.01. On inspection of the genomic sequence, an extra region (#2) was found to have a clustering of four Brk binding motifs within a 350 nt region. (B) All four regions of the Myc genomic region can be activated by Mad/Medea and repressed by Brk. Relative luminescence of a firefly luciferase reporter containing an Hsp70 basal promoter (‘control reporter’) or the same reporter into which four different Myc genomic regions indicated in A were introduced, relative to a renilla control reporter. Where indicated, S2 cells were co-transfected to express activated Thickveins+Mad+Medea (‘+Dpp’) and Brk (‘+Brk’), n=3. (C) Mutation (mut) of the Brk binding sites renders the Myc genomic regions insensitive to Brk. The indicated genomic sequences were trimerized and cloned into a luciferase reporter as in B. Brk binding motifs (GGCGYY) or Mad binding motifs (GRCGNC) are indicated by red and blue arrows, respectively. Point mutagenesis (underlined) removed the Brk binding motif without disturbing the Mad binding motif, n=3. Values for each reporter are normalized to 1 for the ‘−’ condition (B,C). Error bars: standard deviation. *t-test ⩽0.05, **t-test ⩽0.01, ***t-test ⩽0.001. Brk, Brinker; Chr, chromosome; ChIP-Seq, chromatin immunoprecipitation sequence; NS, nonsignificant; WT, wild-type.