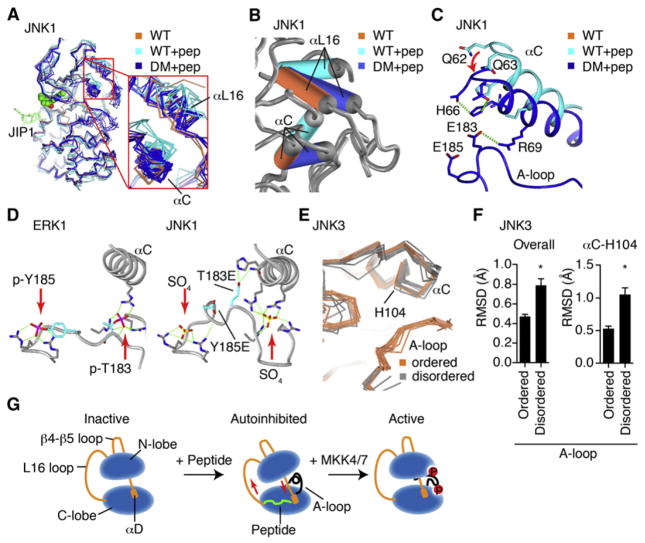
Figure 4. Activation Loop Control of Inter-lobe Rotation.
(A) All of the published JNK1 structures (Table S1C) were superimposed and shown as α-carbon trace, including wild-type (2 structures, coral), wild-type with JIP1 peptide (2 structures, cyan), and mutants of the phosphoacceptor residues in the A-loop to Glu (12 structures, blue).
(B) One JNK1 structure in each class was superimposed, showing how the A-loop mutations partially restore the peptide-induced interlobe rotation of αC and αL16.
(C) T183E in the mutant JNK1 structure H-bonds with residues in αC, drawing it closer to the wild-type, peptide-free positioning.
(D) Comparison of phosphoERK1 structure (2ERK. pdb) with phosphoacceptor mutant JNK1 (3O2M. pdb) structure, showing high structural homology, and suggesting that the mutant T183E likely binds differently to αC than phospho-T183 would.
(E) The JNK3 structures with inhibitors and no peptide were superimposed against 1JNK.pdb, of which 12 show an ordered A-loop, and 12 do not. Show is how the ordered A-loop enforces a single common conformer of αC.
(F) Backbone RMSD from 1JNK.pdb was calculated for the structures superimposed in (D). The RMSD for α-carbon in αC His104 was also calculated. *p < 0.0001 using Student’s t test for significance. Error bars represent SEM.
(G) Model of peptide-induced autoinhibition, and restoration of the active conformer by phosphorylation of the active loop.
See also Figure S4.