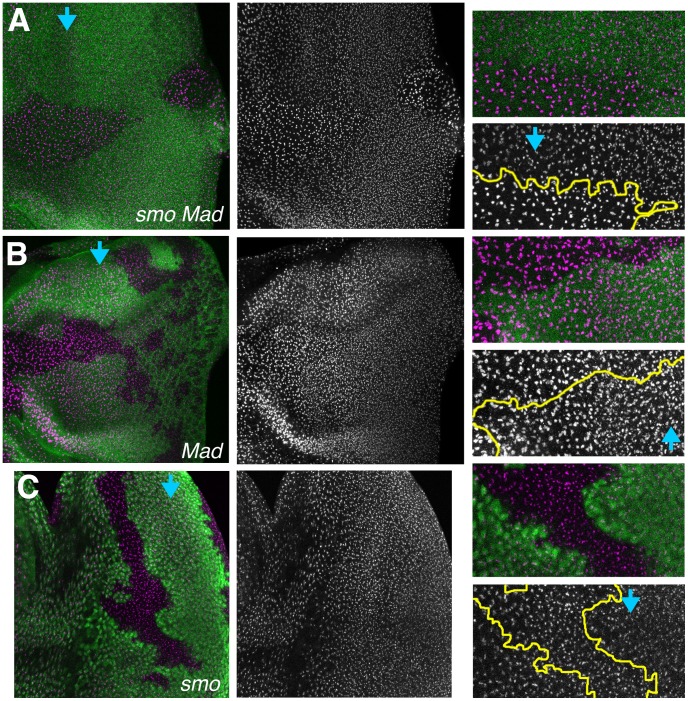
Figure 7. Requirement for Dpp and Hh signaling in nucleolar size.
A. Eye imaginal disc labelled for nucleoli (magenta). Clones mutant for smo and Mad lack beta-galactosidase expression (green). Blue arrow shows the onset of fate specification in the morphogenetic furrow. Nucleoli remain the same size within smo Mad clones. Outside the clone nucleoli become progressively smaller. Nucleoli mutant for smo and Mad also appear more intensely labelled. Fibrillarin channel and enlargements shown to the right. The distinction is evident posterior to the furrow in the enlarged panels. B. Clones mutant for Mad lack beta-galactosidase expression (green). Nucleoli mutant for Mad shrink at the same time as in wild type cells. Fibrillarin channel and enlargements shown to the right. C. Clones mutant for smo lack beta-galactosidase expression (green). Nucleoli mutant for smo shrink at the same time as in wild type cells. Fibrillarin channel and enlargements shown to the right. In panels A–C, fibrillarin labeling is shown as a maximum projection of multiple layers, because nucleoli occupy varying positions in the z-axis. Beta-galactosidase labeling is shown for only a single, central, z-plane, so that parts of the negatively-marked clones were not obscured. To accurately determine the genotype of each nucleolus, beta-galactosidase and fibrillarin labeling must be examined for each confocal plane (not shown).