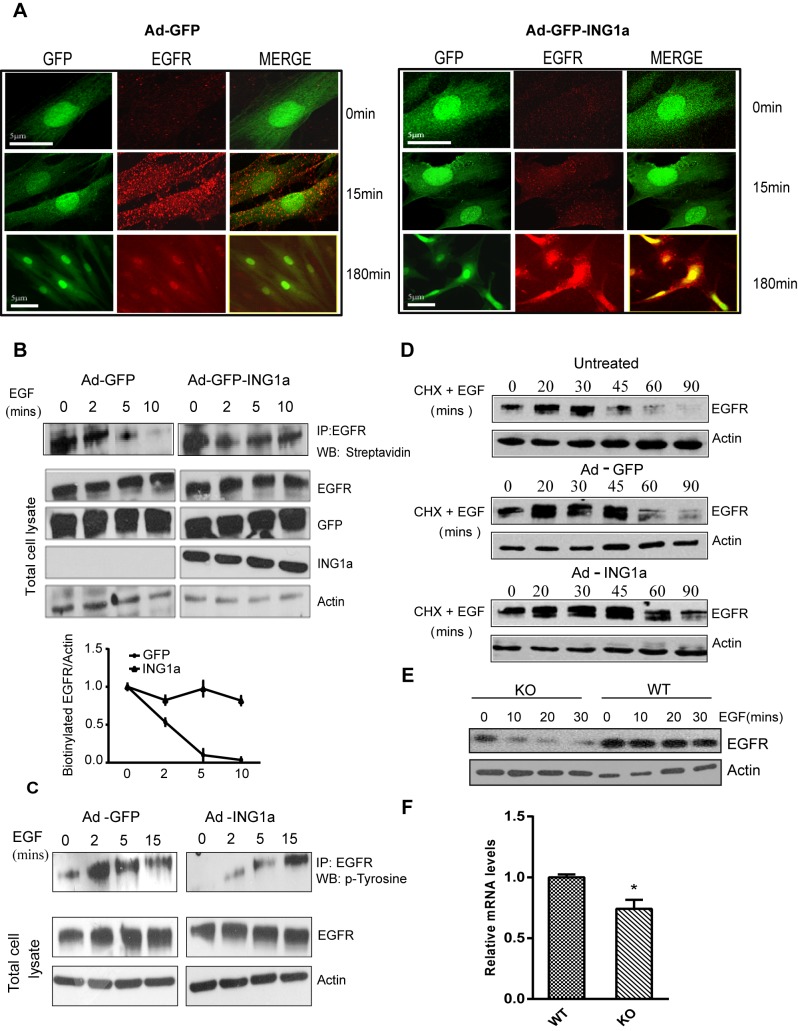
Figure 2. Regulation of endocytosis by ING1a.
(A) Cells infected with adenovirus-GFP or adenovirus-GFP-ING1a (with GFP and ING1a expressed under separate promoters) were serum-deprived for 12 h, stimulated with 100 ng/ml EGF, and fixed 15 or 180 min later. Indirect immunofluorescence for EGFR showed reduced amounts of internalized EGFR in cells expressing ING1a at early time points but persistence of the EGFR at later times. All cells were treated with 10 µg/ml of cycloheximide to inhibit de novo protein synthesis. We took 0- and 15-min images using 63× objectives, while 180 min were imaged at 40× magnification. (B) Biotin internalization assay. Ad-GFP-ING1a and Ad-GFP-expressing cells were serum-starved and EGF-stimulated for the indicated times. Total cell surface proteins were biotinylated, and EGFR was immunoprecipiatated. Biotinylated cell surface EGFR at the indicated time points was detected using streptavidin-HRP. The graph shows the rate of EGFR internalization in GFP- and ING1a-expressing cells as estimated by scanning densitometry. (C) A431 cells infected with adenovirus-GFP/ING1a were serum-starved and checked for tyrosine phosphorylation of the immunoprecipiated EGFR upon EGF stimulation for the indicated time-points. (D) Cells left untreated or expressing GFP or GFP plus ING1a for 48 h were serum-starved overnight (12 h) and stimulated with 100 ng/ml EGF for the indicated times. 100 µg/ml of cycloheximide was used to inhibit protein synthesis. Cells were lysed and levels of EGFR were analyzed by western blotting. Actin was used as a loading control. (E) Wild-type or ING1 knockout MEFs were serum-starved overnight, treated with 10 µg/ml of cycloheximide for 20 min, stimulated with 100 ng/ml EGF and were harvested at the indicated times. Proteins were resolved by SDS-PAGE and blotted with α-EGFR antibody and α-actin as loading control. (F) mRNA levels of Ese2, the ITSN2 mouse homologue in MEF WT and ING−/− cells. RNA was isolated from these cells, and qRT-PCR was performed on three independent replicates. The gene expression levels were normalized to GAPDH (p<0.05).