Figure 2. Principles underlying PPR-RNA recognition.
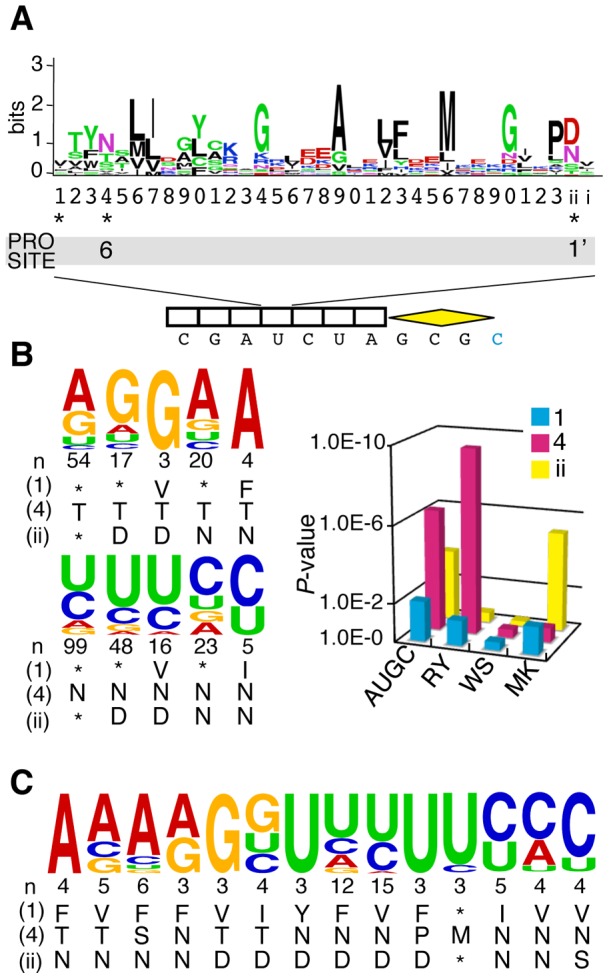
(A) Consensus amino acids for the PPR motif. The sequence logo was derived from 5668 individual PPR motifs (PROSITE accession no. PS51375). Residues 1, 4, and “ii” (position -2) were determined to be nucleotide-specifying residues (NSRs; asterisks). Previously identified residues for RNA recognition (6 and 1′, [20]) are also shown according to the PROSITE model. The last PPR motif was located 4 nucleotides before the editable C residue (Aln4 in Figure 1). (B) NSR-deduced nucleotide frequencies. Nucleotide frequencies, determined according to the NSRs, are displayed in a logo (left panel). * indicates any amino acid; “n” indicates the occurrence frequencies of the NSRs in 327 Arabidopsis PRR-motifs. The nucleotide-specifying capacity of a residue was deduced from the low variability in the association between the individual amino acids and nucleotides and is presented as a P-value (right panel). The analysis was conducted for specific nucleotides (i.e., A, U, G, or C), purine/pyrimidine (R or Y), presence or absence of hydrogen bond groups (W or S), and presence or absence of amino/keto (M or K) groups, in alignment 4 (Aln4). (C) Example for deduced nucleotide frequencies by various combinations of NSRs.
