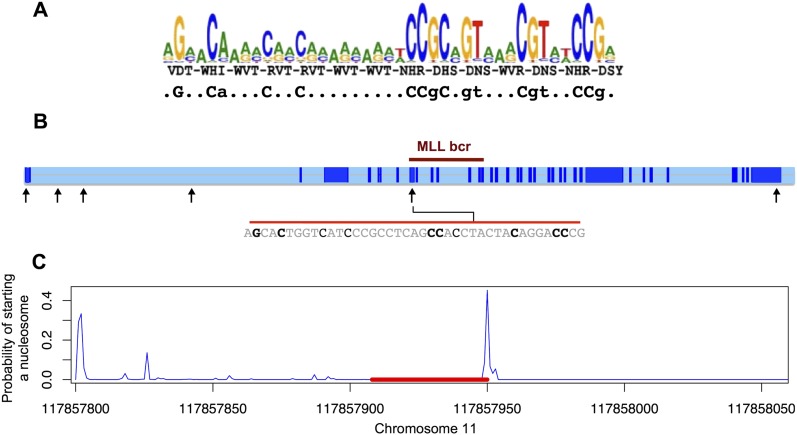
Figure 4.
PRDM9 C binding motif in the MLL breakpoint cluster region. (A) Logo plot of the C allele binding motif (Baudat et al. 2010), predicted based on the three indicated residues forming the binding unit of the ZnF repeats (positions −1, 3, and 6 of the ZnF alpha helices) and the consensus sequence motif simplified showing the most strongly predicted bases (in lowercase for >80% consensus for a specific base and in uppercase for >95% consensus; Berg et al. 2010). (B) Presence of a motif at chr11:117857908–117857950 (hg18), within the breakpoint cluster region of MLL, matching the predicted PRDM9 C allele binding motif for the seven strongly predicted bases shown in uppercase in the consensus sequence presented in A, and three predicted bases shown in lowercase. (Light blue) Intronic regions. (Black arrows) Positions of all occurrences of sequences matching the motif at uppercase characters. No PRDM9 A allele binding motif was found in MLL. (C) Nucleosome starting positions predicted by NuPoP (Xi et al. 2010). (Red line) Position of the predicted C binding motif.