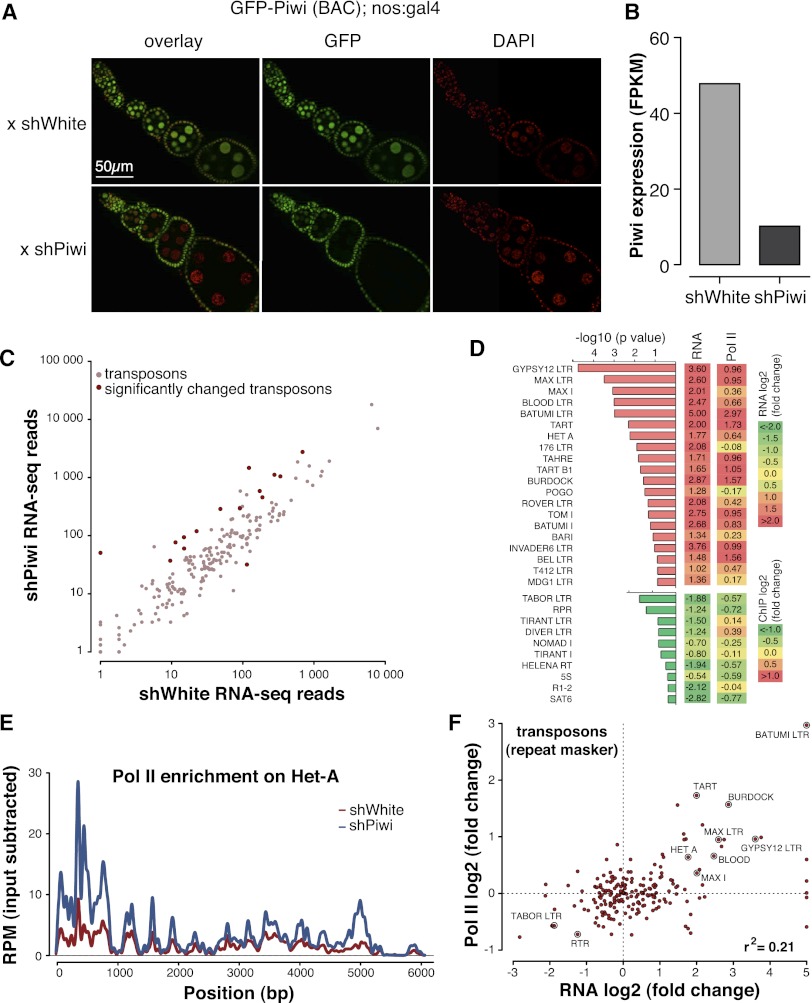
Figure 3.
Piwi transcriptionally represses TEs. (A) Piwi knockdown is efficient and specific to ovarian germ cells as indicated by GFP-Piwi localization. GFP-Piwi; Nanos-Gal4-VP16 flies were crossed to control shRNA (shWhite) or shPiwi lines. Piwi is specifically depleted in germ cells and not in follicular cells, consistent with expression of the Nanos-Gal4-VP16 driver. (B) Piwi expression as measured by RNA-seq in the Piwi knockdown and control lines. Note that Piwi expression is unaffected in follicular cells, leading to relatively weak apparent knockdown in RNA-seq libraries from whole ovaries. (C) Effect of Piwi knockdown on the expression of TEs. Two biological replicate RNA-seq experiments were carried out, and differential expression was assessed using DESeq. Transposons that show significant change (P < 0.05) are indicated by dark-red circles. Out of 217 individual RepeatMasker-annotated TEs, 15 show a significant increase in expression upon Piwi knockdown. (D) The change in the levels of TE transcripts and Pol II occupancy on their promoters upon Piwi knockdown. Twenty up-regulated and 10 down-regulated transposons with the most significant changes in expression level are shown. Note the low statistical significance for down-regulated transposons. For a complete list of transposons, see Supplemental Figure S2. (E) Pol II signal over the Het-A retrotransposon in control flies (shWhite; red) and upon Piwi knockdown (shPiwi; blue). (F) Increased abundance of transposon transcripts upon Piwi depletion correlates with increased Pol II occupancy over their promoters (r2 = 0.21). Note that the majority of elements do not show significant change in either RNA abundance or Pol II occupancy.