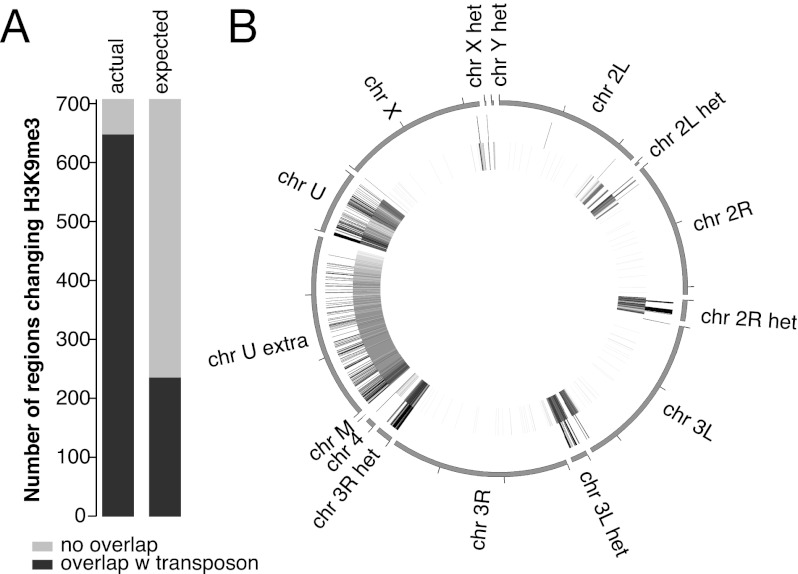
Figure 4.
Piwi-induced transcriptional repression correlates with establishment of a repressive chromatin state. (A) Overlap between genomic regions of H3K9me3 depletion upon Piwi knockdown and TEs. Two replicates of H3K9me3 ChIP-seq experiments were carried out on control and Piwi-depleted ovaries, and enriched regions were identified using DESeq (see the Materials and Methods for details). A total of 705 regions show significant (P < 0.05) decrease in H3K9me3 occupancy upon Piwi knockdown, while only 30 regions showed a similarly significant increase. Out of the 705 regions that show a decrease in H3K9me3 marks upon Piwi knockdown, 91% (646) overlap with TE sequences compared with the 33% expected from random genome sampling. (B) Genomic positions of H3K9me3-depleted regions upon Piwi depletion (outer circle) and RepeatMasker-annotated transposons (inner circle). Note that almost all regions are localized in heterochromatic and repeat-rich portions of the genome (Het, chrU, and chrUExtra chromosomes).