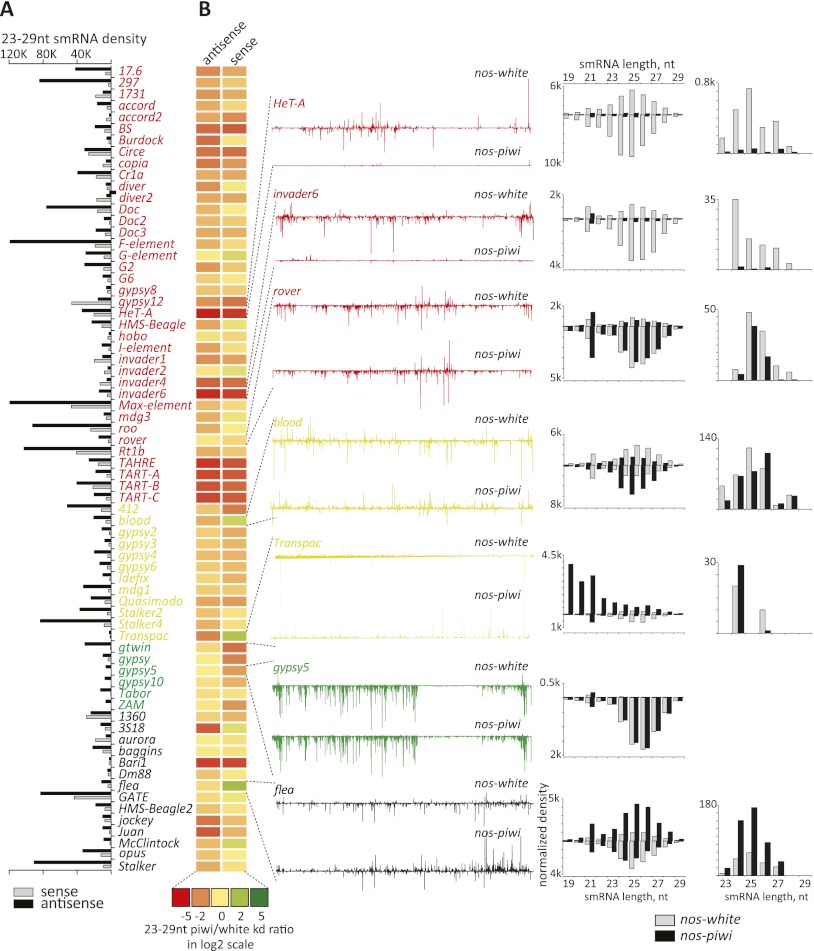
Figure 4.
Impact of germline piwi knockdown on piRNAs mapping to transposons. (A) A bar graph represents the density of piRNA (RPM, strand as indicated) mapping to the 70 most highly targeted elements. These are sorted according to their cell type bias, with the coloration of the names as described in Figure 1. (Red) Germline-biased; (yellow) intermediate; (green) soma-biased; (black) undetermined. (B) A heat map represents changes in small RNAs (scale below) in comparisons of germline piwi knockdown versus control animals. To the left, densities of piRNAs along lines representing the extent of the transposon consensus are shown for the control (top) and piwi knockdown libraries (bottom) for selected elements (indicated). Adjacent to that are size distributions for small RNAs corresponding to those elements, with sense species displayed above the axis and antisense displayed below the axis (piwi knockdown and control as indicated). At the extreme right are similar plots of only that subset of piRNAs (23–29 nt) that is most probably bound to Ago3; namely; those sense to elements that lack a 5′ U and have an A at position 10.