Figure 8.
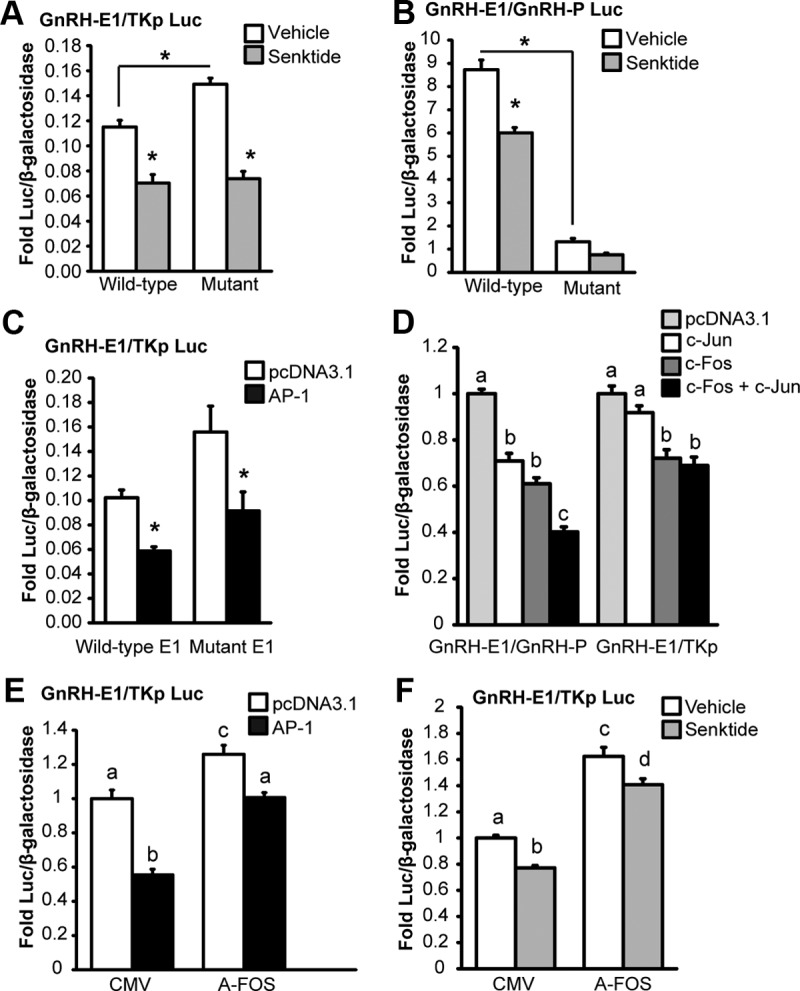
The roles of AP-1 half-sites and AP-1 complex formation in the repression of enhancer 1 and the promoter activity by senktide. Mutation of AP-1 half-sites in enhancer 1 and the promoter of GnRH changes basal transcription but does not block repression by senktide. A, Site-directed mutagenesis was employed to make mutations in the GnRH-E1/TKp Luc reporter that correspond to those shown to disrupt TPA-mediated complex formation on E1A (-1737/−1715) by gel shift. GT1-7 cells were transiently transfected with rat NK3R and a GnRH-E1/TKp Luc reporter with either a wild-type or a mutant AP-1 element and treated for 8 hours with either vehicle or 30 nM senktide. B, Site-directed mutagenesis was employed to make mutations in the GnRH-E1/GnRH-P Luc reporter corresponding to those shown to disrupt TPA-complex formation on PA (-106/−84) by gel shift (E1 is wild-type). GT1-7 cells were transiently transfected with rat NK3R and a GnRH-E1/P Luc reporter with either a wild-type or a mutant AP-1 element and treated for 8 hours with either vehicle or 30 nM senktide. C, GT1-7 cells were transiently transfected with rat NK3R and either a wild-type or a mutant AP-1 GnRH-E1/TKp Luc reporter with AP-1 transcription factors (c-Fos and c-Jun) or empty vector (pcDNA3.1). D, GT1-7 cells were transiently transfected with rat NK3R and either a GnRH-E1/GnRH-P or GnRH-E1/TKp luciferase reporter with empty vector control (pcDNA3.1), c-Fos, or c-Jun individually balanced with pcDNA3.1, or c-Fos and c-Jun in combination. E, GT1-7 cells were transiently transfected with rat NK3R and either A-FOS or CMV (empty vector control) and AP-1 (c-Jun and c-Fos) or pcDNA3.1 (empty vector control). F, C, GT1-7 cells were transiently transfected with rat NK3R and either A-FOS or CMV (empty vector control) and treated for 8 hours with either vehicle or 30 nM senktide. For A through F, luciferase values were internally normalized using a cotransfected β-galactosidase reporter to control for transfection efficiency, and data were normalized to an external control. Data are presented as mean fold ± SEM for n = 3 independent experiments. Comparisons were made using ANOVA with Least squares means Tukey HSD (α = .05). *Significantly different from vehicle or wild-type control (indicated by bracket) or levels not connected by same letter are significantly different.
