Abstract
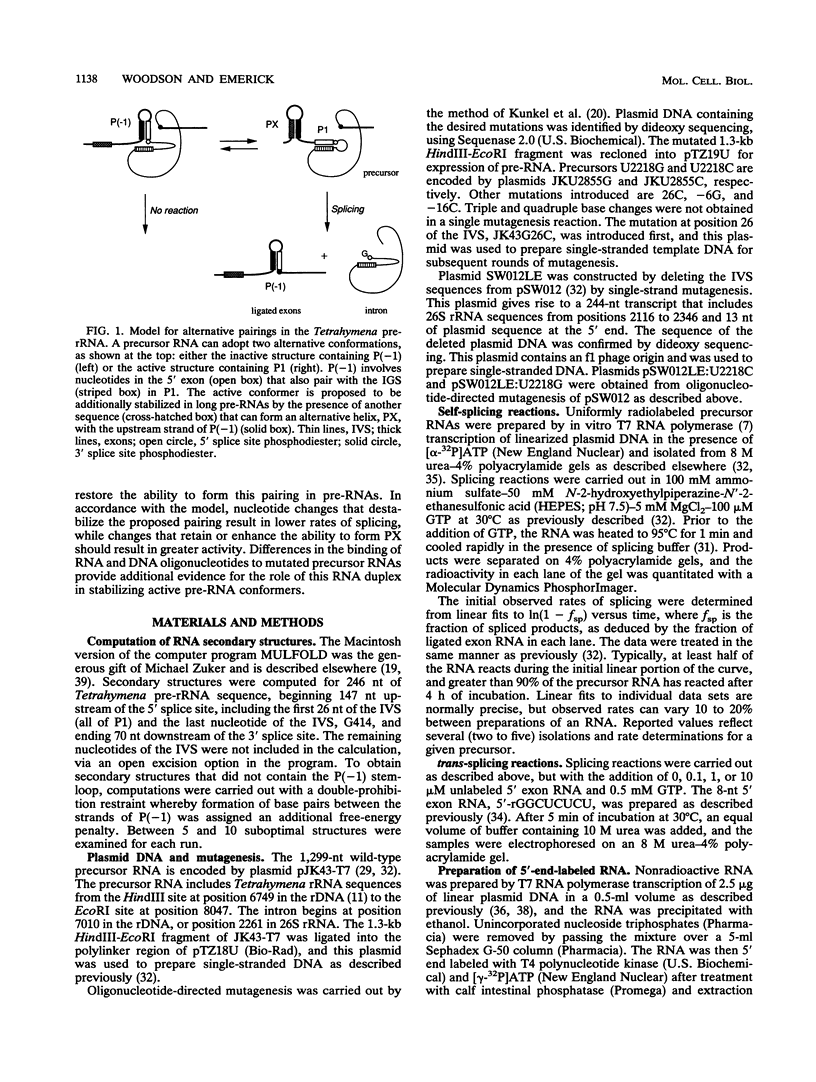
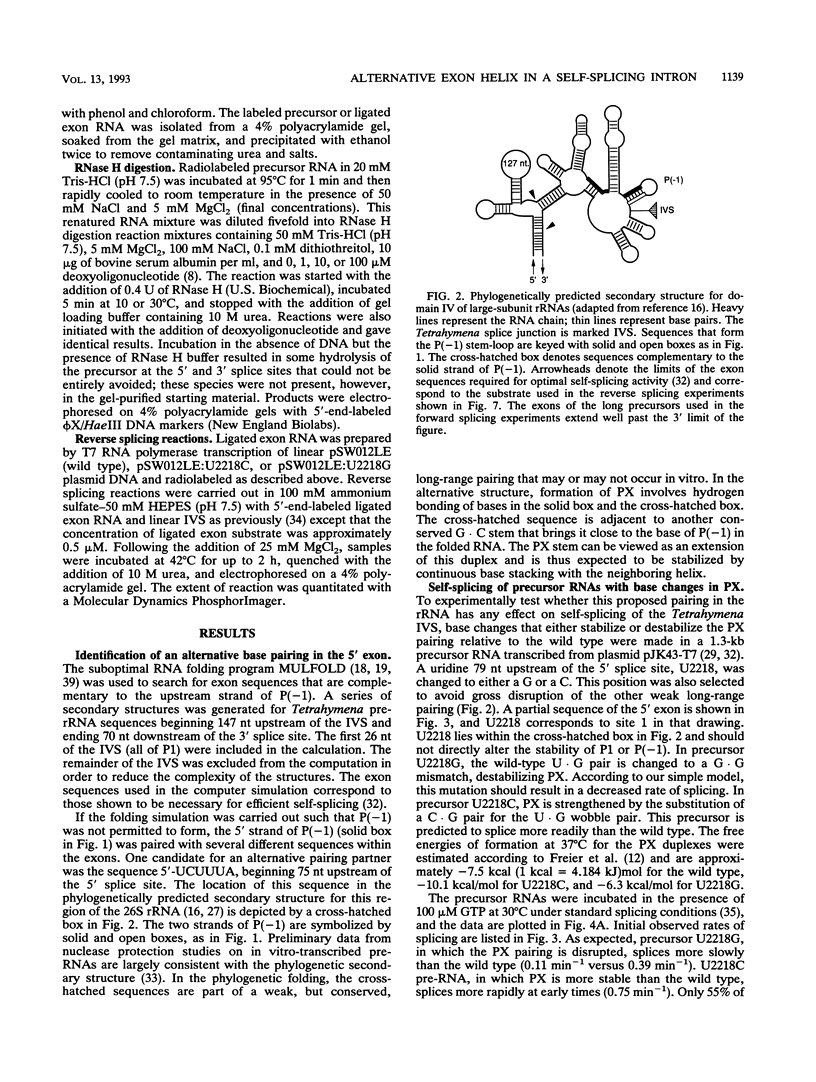
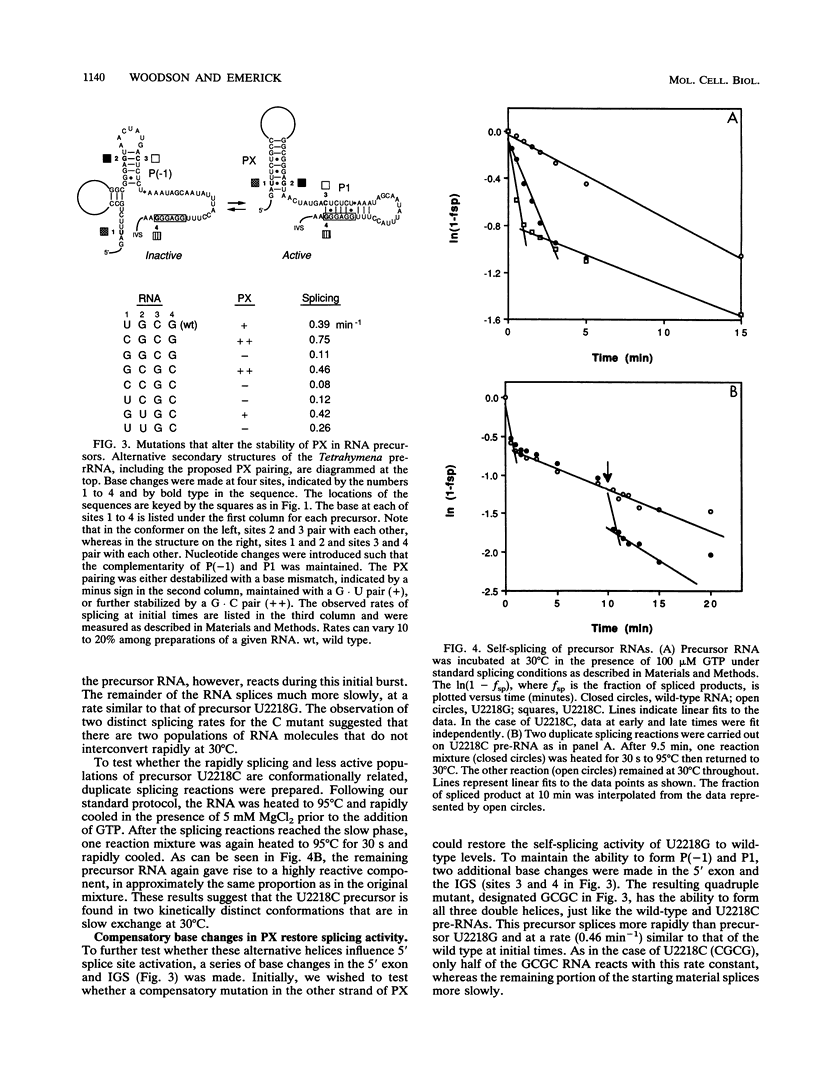
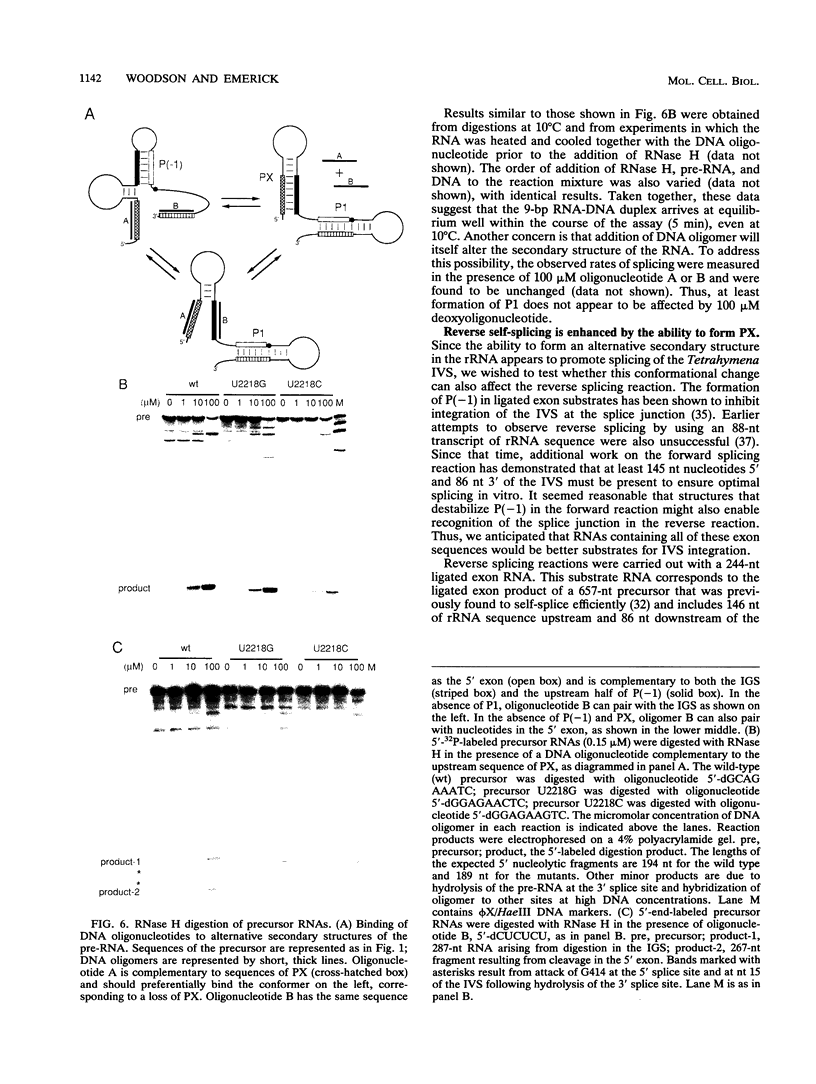
A highly conserved ribosomal stem-loop immediately upstream of the Tetrahymena splice junction can inhibit both forward and reverse self-splicing by competing with base pairing between the 5' exon and the guide sequence of the intervening sequence. Formation of this unproductive hairpin is preferred in precursor RNAs with short exons and results in a lower rate of splicing. Inhibition of self-splicing is not observed in longer precursors, suggesting that additional interactions in the extended exons can influence the equilibrium between the productive and unproductive hairpins at the 5' splice site. An alternative pairing upstream of the 5' splice site has been identified and is proposed to stabilize the active conformer of the pre-rRNA. Nucleotide changes that alter the ability to form this additional helix were made, and the self-splicing rates were compared. Precursors in which the proposed stem is stabilized splice more rapidly than the wild type, whereas RNAs that contain a base mismatch splice more slowly. The ability of DNA oligomers to bind the RNA, as detected by RNase H digestion, correlates with the predicted secondary structure of the RNA. We also show that a 236-nucleotide RNA containing the natural splice junction is a substrate for intervening sequence integration. As in the forward reaction, reverse splicing is enhanced in ligated exon substrates in which the alternative rRNA pairing is more stable.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altuvia S., Kornitzer D., Teff D., Oppenheim A. B. Alternative mRNA structures of the cIII gene of bacteriophage lambda determine the rate of its translation initiation. J Mol Biol. 1989 Nov 20;210(2):265–280. doi: 10.1016/0022-2836(89)90329-x. [DOI] [PubMed] [Google Scholar]
- Bass B. L., Cech T. R. Specific interaction between the self-splicing RNA of Tetrahymena and its guanosine substrate: implications for biological catalysis by RNA. 1984 Apr 26-May 2Nature. 308(5962):820–826. doi: 10.1038/308820a0. [DOI] [PubMed] [Google Scholar]
- Been M. D., Cech T. R. One binding site determines sequence specificity of Tetrahymena pre-rRNA self-splicing, trans-splicing, and RNA enzyme activity. Cell. 1986 Oct 24;47(2):207–216. doi: 10.1016/0092-8674(86)90443-5. [DOI] [PubMed] [Google Scholar]
- Cech T. R. Self-splicing RNA: implications for evolution. Int Rev Cytol. 1985;93:3–22. doi: 10.1016/s0074-7696(08)61370-4. [DOI] [PubMed] [Google Scholar]
- Cech T. R. Self-splicing of group I introns. Annu Rev Biochem. 1990;59:543–568. doi: 10.1146/annurev.bi.59.070190.002551. [DOI] [PubMed] [Google Scholar]
- Chebli K., Gattoni R., Schmitt P., Hildwein G., Stevenin J. The 216-nucleotide intron of the E1A pre-mRNA contains a hairpin structure that permits utilization of unusually distant branch acceptors. Mol Cell Biol. 1989 Nov;9(11):4852–4861. doi: 10.1128/mcb.9.11.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouet d'Orval B., d'Aubenton Carafa Y., Sirand-Pugnet P., Gallego M., Brody E., Marie J. RNA secondary structure repression of a muscle-specific exon in HeLa cell nuclear extracts. Science. 1991 Jun 28;252(5014):1823–1828. doi: 10.1126/science.2063195. [DOI] [PubMed] [Google Scholar]
- Davanloo P., Rosenberg A. H., Dunn J. J., Studier F. W. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2035–2039. doi: 10.1073/pnas.81.7.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donis-Keller H. Site specific enzymatic cleavage of RNA. Nucleic Acids Res. 1979 Sep 11;7(1):179–192. doi: 10.1093/nar/7.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng F. J., Warner J. R. Structural basis for the regulation of splicing of a yeast messenger RNA. Cell. 1991 May 31;65(5):797–804. doi: 10.1016/0092-8674(91)90387-e. [DOI] [PubMed] [Google Scholar]
- Engberg J., Nielsen H. Complete sequence of the extrachromosomal rDNA molecule from the ciliate Tetrahymena thermophila strain B1868VII. Nucleic Acids Res. 1990 Dec 11;18(23):6915–6919. doi: 10.1093/nar/18.23.6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M. R. Biochemical mechanisms of constitutive and regulated pre-mRNA splicing. Annu Rev Cell Biol. 1991;7:559–599. doi: 10.1146/annurev.cb.07.110191.003015. [DOI] [PubMed] [Google Scholar]
- Gutell R. R., Schnare M. N., Gray M. W. A compilation of large subunit (23S-like) ribosomal RNA sequences presented in a secondary structure format. Nucleic Acids Res. 1990 Apr 25;18 (Suppl):2319–2330. doi: 10.1093/nar/18.suppl.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T., Sullivan F. X., Cech T. R. Intermolecular exon ligation of the rRNA precursor of Tetrahymena: oligonucleotides can function as 5' exons. Cell. 1985 Dec;43(2 Pt 1):431–437. doi: 10.1016/0092-8674(85)90173-4. [DOI] [PubMed] [Google Scholar]
- Jaeger J. A., Turner D. H., Zuker M. Improved predictions of secondary structures for RNA. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger J. A., Turner D. H., Zuker M. Predicting optimal and suboptimal secondary structure for RNA. Methods Enzymol. 1990;183:281–306. doi: 10.1016/0076-6879(90)83019-6. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Libri D., Piseri A., Fiszman M. Y. Tissue-specific splicing in vivo of the beta-tropomyosin gene: dependence on an RNA secondary structure. Science. 1991 Jun 28;252(5014):1842–1845. doi: 10.1126/science.2063196. [DOI] [PubMed] [Google Scholar]
- Maniatis T. Mechanisms of alternative pre-mRNA splicing. Science. 1991 Jan 4;251(4989):33–34. doi: 10.1126/science.1824726. [DOI] [PubMed] [Google Scholar]
- Marconi R. T., Hill W. E. Evidence for a tRNA/rRNA interaction site within the peptidyltransferase center of the Escherichia coli ribosome. Biochemistry. 1989 Jan 24;28(2):893–899. doi: 10.1021/bi00428a073. [DOI] [PubMed] [Google Scholar]
- Mohr G., Lambowitz A. M. Integration of a group I intron into a ribosomal RNA sequence promoted by a tyrosyl-tRNA synthetase. Nature. 1991 Nov 14;354(6349):164–167. doi: 10.1038/354164a0. [DOI] [PubMed] [Google Scholar]
- Murphy F. L., Cech T. R. Alteration of substrate specificity for the endoribonucleolytic cleavage of RNA by the Tetrahymena ribozyme. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9218–9222. doi: 10.1073/pnas.86.23.9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller H. F. Ribosomal RNA and translation. Annu Rev Biochem. 1991;60:191–227. doi: 10.1146/annurev.bi.60.070191.001203. [DOI] [PubMed] [Google Scholar]
- Noller H. F. Structure of ribosomal RNA. Annu Rev Biochem. 1984;53:119–162. doi: 10.1146/annurev.bi.53.070184.001003. [DOI] [PubMed] [Google Scholar]
- Price J. V., Kieft G. L., Kent J. R., Sievers E. L., Cech T. R. Sequence requirements for self-splicing of the Tetrahymena thermophila pre-ribosomal RNA. Nucleic Acids Res. 1985 Mar 25;13(6):1871–1889. doi: 10.1093/nar/13.6.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. A. On the origin of RNA splicing and introns. Cell. 1985 Sep;42(2):397–400. doi: 10.1016/0092-8674(85)90092-3. [DOI] [PubMed] [Google Scholar]
- Walstrum S. A., Uhlenbeck O. C. The self-splicing RNA of Tetrahymena is trapped in a less active conformation by gel purification. Biochemistry. 1990 Nov 20;29(46):10573–10576. doi: 10.1021/bi00498a022. [DOI] [PubMed] [Google Scholar]
- Woodson S. A., Cech T. R. Alternative secondary structures in the 5' exon affect both forward and reverse self-splicing of the Tetrahymena intervening sequence RNA. Biochemistry. 1991 Feb 26;30(8):2042–2050. doi: 10.1021/bi00222a006. [DOI] [PubMed] [Google Scholar]
- Woodson S. A., Cech T. R. Reverse self-splicing of the tetrahymena group I intron: implication for the directionality of splicing and for intron transposition. Cell. 1989 Apr 21;57(2):335–345. doi: 10.1016/0092-8674(89)90971-9. [DOI] [PubMed] [Google Scholar]
- Woodson S. A. Exon sequences distant from the splice junction are required for efficient self-splicing of the Tetrahymena IVS. Nucleic Acids Res. 1992 Aug 11;20(15):4027–4032. doi: 10.1093/nar/20.15.4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaug A. J., Been M. D., Cech T. R. The Tetrahymena ribozyme acts like an RNA restriction endonuclease. Nature. 1986 Dec 4;324(6096):429–433. doi: 10.1038/324429a0. [DOI] [PubMed] [Google Scholar]
- Zaug A. J., Cech T. R. Oligomerization of intervening sequence RNA molecules in the absence of proteins. Science. 1985 Sep 13;229(4718):1060–1064. doi: 10.1126/science.2412290. [DOI] [PubMed] [Google Scholar]
- Zaug A. J., Cech T. R. The intervening sequence RNA of Tetrahymena is an enzyme. Science. 1986 Jan 31;231(4737):470–475. doi: 10.1126/science.3941911. [DOI] [PubMed] [Google Scholar]
- Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989 Apr 7;244(4900):48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]