Abstract
Purpose
Previously, we showed that microRNA (miRNA) signatures derived from the peripheral blood of mice are highly specific for both radiation energy (γ-rays or high linear energy transfer [LET] 56Fe ions) and radiation dose. Here, we investigate to what extent miRNA expression signatures derived from mouse blood can be used as biomarkers for exposure to 600 MeV proton radiation.
Materials and methods
We exposed mice to 600 MeV protons, using doses of 0.5 or 1.0 Gy, isolated total RNA at 6 h or 24 h after irradiation, and used quantitative real-time polymerase chain reaction (PCR) to determine the changes in miRNA expression.
Results
A total of 26 miRNA were differentially expressed after proton irradiation, in either one (77%) or multiple conditions (23%). Statistical classifiers based on proton, γ, and 56Fe-ion miRNA expression signatures predicted radiation type and proton dose with accuracies of 81% and 88%, respectively. Importantly, gene ontology analysis for proton-irradiated cells shows that genes targeted by radiation-induced miRNA are involved in biological processes and molecular functions similar to those controlled by miRNA in γ ray- and 56Fe-irradiated cells.
Conclusions
Mouse blood miRNA signatures induced by proton, γ, or 56Fe irradiation are radiation type- and dose-specific. These findings underline the complexity of the miRNA-mediated radiation response.
Keywords: miRNA, protons, biodosimetry, high-LET radiation, class prediction, gene ontology analysis
Introduction
MicroRNA (miRNA) are a class of small non-coding RNA that are important regulators of gene expression (Bartel 2009). In vivo and in vitro studies have found that miRNA control is essential for the proper execution of many processes in normal cells, including cell metabolism, cell differentiation, and cell signaling (Maes et al. 2008, Shkumatava et al. 2009). A host of recent findings demonstrate that radiation has a significant impact on miRNA expression (reviewed in Dickey et al. 2011). Among these studies are several examples of low linear energy transfer (LET) effects on normal human fibroblasts and various immortalized cell lines (Ishii and Saito 2006, Marsit et al. 2006, Josson et al. 2008, Maes et al. 2008, Chaudhry 2009, Shin et al. 2009, Simone et al. 2009, Nikiforova et al. 2011). The exact role of miRNA in radiation response biology is unclear, and the mechanisms for its participation in post-radiation events remain to be determined. Intriguingly, miRNA has a role in both direct or indirect effects of radiation, as was shown in higher-order organoid cultures (Dickey et al. 2011). miRNA induction by either low-LET X-ray or high-LET 56Fe-ion radiation may also play a role in chromatin remodeling and DNA methylation (Aypar et al. 2011). A role for miRNA in pro-survival capacity after irradiation is supported by findings in an endothelium-derived cell line (Kraemer et al. 2011). It was also shown that plasma miRNA signatures are dose dependent (Josson et al. 2008). We have reported a strong effect of radiation on miRNA expression profiles derived from the circulating blood of radiotherapy patients (Templin et al. 2011b). We also showed that blood miRNA expression signatures of mice exposed to low- or high-LET radiation (γ-rays or 56Fe ions) are highly dependent on radiation energy and dose (Templin et al. 2011a). Classifiers based on these signatures reliably predicted the irradiation type and dose of mice with unknown irradiation status. Together, these results demonstrate that miRNA signatures may be used as both radiation biodosimeters and indicators of radiation-induced functional changes in cells and tissues.
In this study, we investigate the potential of miRNA signatures to be used as biomarkers for exposure to 600 MeV protons. Protons are the main type of high-LET radiation encountered by astronauts in low earth orbit. They are components of the solar wind, the Van Allen radiation belts, and galactic cosmic rays and cumulatively pose a health threat to astronauts, particularly during heightened solar activity. In this in vivo proof-of-principle study, we exposed C57BL/6 mice to 600 MeV proton total body irradiation and measured miRNA expression levels in the blood of the irradiated and control animals. Using these results and the results that we obtained from prior work (Templin et al. 2011a), we developed statistical classifiers that can be used in the estimation of exposure parameters. Finally, we analyzed the involvement of the differentially expressed miRNA in cellular processes.
Materials and methods
Animals and irradiation
Eighteen male C57BL/6 mice were obtained from Taconic (Hudson, NY, USA). Animals were kept in holding cages in groups of ~ 3, on a regular 12 h/12 h light/dark cycle with ad libitum access to medium-fat standard lab chow and water. Animals were 12 weeks of age when they were exposed to radiation. Three animals were used per condition, except for the 6 h post sham-irradiation control condition, for which only two animals were available. The mice were exposed to doses of 0 (sham-irradiated control), 0.5, or 1.0 Gy 600 MeV protons at a dose rate of 0.2 Gy/min at the National Aeronautics and Space Administration (NASA) Space Radiation Laboratory of Brookhaven National Laboratory. Extra care was taken to avoid stressing the animals during the experiments, in order to minimize stress-induced changes in gene expression and to ensure that the measured miRNA expression changes were indeed caused by irradiation. For this reason, we irradiated the mice in relatively large cages that did not unnecessarily restrain the animals’ movements. Mice were individually irradiated in boxes sized 11 × 8 × 6 cm (w/l/h), which were kept in the mouse cages for 24 h before irradiation. Animals that did not receive the maximum radiation dose were kept in their boxes in the radiation facility for the same time as the animals exposed to the highest dose (5 min) in order to standardize treatment conditions. All animal husbandry and experimental procedures were conducted in accordance with applicable federal and state guidelines and approved by the Animal Care and Use Committees of Columbia University Medical Center and Brookhaven National Laboratory.
Blood collection and RNA isolation
Six or 24 h after irradiation, 250 µl blood was collected from the mice using the submandibular method. The blood was collected directly in tubes containing lysis solution, a procedure that preserves the in vivo miRNA expression signatures. Three samples were collected per condition, resulting in a sample size of n = 3 biological repeats per irradiation dose and time point, with the exception of the 6-h post sham-irradiation control condition, which had a sample size of n = 2. Each mouse was bled only once, and different animals were used for each irradiation condition (time point and dose). Total RNA was purified using the mirVana™ PARIS™ kit according to the manufacturer’s instructions (Life Technologies, Carlsbad, CA, USA). Neither blood nor RNA from different animals was pooled, and all samples were processed and analyzed separately. RNA concentration was measured on a Nanodrop 1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA), and RNA integrity was determined using the Agilent 2100 Bioanalyzer microelectrophoretic system (Agilent Technologies, Santa Clara, CA, USA).
Determination of miRNA expression signatures by means of quantitative PCR
In order to determine miRNA expression levels, the miRNA contained in samples of 50 ng total RNA were reverse-transcribed into cDNA using Megaplex™ miRNA-specific stem-loop primers and MultiScribe™ reverse transcriptase. Complementary DNA were preamplified in 12 cycles and then amplified for 40 cycles in 384-well low-density TaqMan® rodent miRNA expression arrays for quantitative real-time polymerase chain reaction (PCR) (all Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions. This methodology has been shown to provide data with greater precision than data obtained from microarrays, which are based on solid-surface hybridizations between oligonucleotide probes and target molecules, and this technology is routinely used to validate data obtained from microarray studies (Pradervand et al. 2010, Yauk et al. 2010). Also, the determination of relative miRNA expression by means of quantitative PCR is highly correlated to expression results obtained from RNA sequencing-based techniques (Pradervand et al. 2010).
Data processing for determination of differentially expressed miRNA
The quantitative real-time PCR data was imported into RQ Manager v. 1.2 (Life Technologies, Carlsbad, CA, USA) to determine cycle threshold (CT) values. The data was then exported to Excel (Microsoft Corporation, Redmond, WA, USA) for preprocessing. Preprocessing included the removal of the non-relevant rat miRNA and of the miRNA that did not have detectable expression (CT = 40) in at least one sample in both the irradiated and time-matched control samples. The preprocessed data was then imported into DataAssist v. 2.0 software (Applied Biosystems) for the first part of the statistical analysis. The statistical analysis included three steps: (i) Normalization of the expression data, (ii) calculation of the P-values of the differentially expressed miRNA, and (iii) calculation of the false discovery rate (FDR) for the results from step 2. The average CT values of miRNA miR-7b and miR-200c were used for normalization of the remaining miRNA in the respective irradiation conditions because the CT values of these two miRNA had the smallest standard deviations across all conditions. Three endogenous controls supplied with an array proved to be radiation-sensitive (small nuclear RNA RNU6B and small nucleolar RNAs snoRNA 135 and snoRNA 202). Normalized CT (ΔCT) values were imported into Excel, and, using the Excel add-in BRB-ArrayTools v. 3.8 (Wright and Simon 2003, Simon et al. 2007), a random-variance t-test was used to calculate individual P-values for the miRNA expression levels in the different irradiation conditions, with the respective calibrator control samples serving as the reference group. FDR were calculated according to the method of Benjamini and Hochberg (1995). miRNA with FDR of less than 0.07 were included in the set of differentially expressed miRNA.
Additionally, statistical power was monitored using the methodology of Lee and Whitmore (2002) and Lee (2004), permitting an assessment of the sensitivity of our analyses. This approach assumes that the differential expression values follow an approximate multivariate normal distribution, based on independent log-intensity values derived from multiple repeated observations and the central limit theorem. Our analysis is based on the R package ‘sizepower’ by Qiu, Lee, and Whitmore (2010). The analysis requires specific inputs. The set of inputs includes the mean number of false positives, the anticipated number of unaffected genes, the mean difference in log-expression between treatment and control conditions as postulated under the alternative hypothesis, the anticipated standard deviation of the difference in log-expression between treatment and control conditions, and lastly the group sample size.
Standard errors of the mean (SEM) for fold changes shown in Table I contain the variability of both control and irradiated samples and were calculated according to the following formula:
with SDΔCT depicting the standard deviation (SD) among the ΔCT values of the samples belonging to a group.
Table I.
MicroRNA differentially expressed in whole mouse blood after exposure to 600 MeV protons in vivo. Mice were irradiated with doses of 0.5 or 1.0 Gy, 250 µl blood was collected directly in lysis solution, and total RNA was purified at 6 and 24 h after irradiation. miRNA expression levels were determined by 384-well low-density TaqMan® real-time PCR miRNA expression assays. The expression data was preprocessed and normalized. Fold changes in expression level, relative to non-irradiated control samples, with a false discovery rate of < 0.07 were considered to be statistically significant.
| Irradiation condition |
miRNA ID | Fold changea ± SEM | P-value | FDR | Avg. CT controlsb |
Avg. CT irradiatedb |
|---|---|---|---|---|---|---|
| p 6 h 0.5 Gy | miR-10b | −5,000.0 ± 1.67 | 2.9E-04 | 0.0135 | 27.7 | 40.0 |
| miR-292-3p | −90.9 ± 1.54 | 0.0011 | 0.0426 | 33.5 | 40.0 | |
| miR-495 | −120.5 ± 1.20 | 8.8-05 | 0.0056 | 33.1 | 40.0 | |
| miR-501-3p | 5,970.0 ± 1.23 | 4.2E-05 | 0.0040 | 40.0 | 27.5 | |
| miR-667 | −384.0 ± 1.08 | 7.4E-06 | 0.0014 | 31.4 | 40.0 | |
| p 6 h 1.0 Gy | miR-1 | −4.4 ± 1.29 | 0.0041 | 0.0482 | 24.5 | 26.7 |
| miR-10b | −4,760.0 ± 1.67 | 1.3E-05 | 6.1E-04 | 27.7 | 40.0 | |
| miR-34c | −16.1 ± 1.65 | 0.0012 | 0.0325 | 35.9 | 40.0 | |
| miR-99a | −2.4 ± 1.19 | 0.0074 | 0.0615 | 24.1 | 25.4 | |
| miR-100 | −2.4 ± 1.29 | 0.0098 | 0.0633 | 24.2 | 25.6 | |
| miR-125b-5p | −3.3 ± 1.26 | 0.0019 | 0.0415 | 23.2 | 25.1 | |
| miR-127 | −4.8 ± 1.32 | 0.0035 | 0.0482 | 25.9 | 28.2 | |
| miR-143 | −13,000.0 ± 1.39 | 1.4E-06 | 1.4E-04 | 26.2 | 40.0 | |
| miR-200a | −16,400.9 ± 1.18 | 1.0E-07 | 1.9E-05 | 25.9 | 40.0 | |
| miR-203 | −5.6 ± 1.53 | 0.0118 | 0.0633 | 21.0 | 23.6 | |
| miR-204 | −4.8 ± 1.47 | 0.0130 | 0.0646 | 26.7 | 29.0 | |
| miR-218 | −4.8 ± 1.29 | 0.0025 | 0.0427 | 25.3 | 27.7 | |
| miR-224 | −3.3 ± 1.40 | 0.0082 | 0.0615 | 28.7 | 30.6 | |
| miR-294 | 34.0 ± 1.32 | 1.6E-04 | 0.0050 | 40.0 | 35.0 | |
| miR-379 | −6.7 ± 1.39 | 0.0029 | 0.0427 | 28.5 | 31.4 | |
| miR-380-5p | −52.6 ± 2.79 | 0.0046 | 0.0495 | 34.2 | 40.0 | |
| miR-411 | −7.1 ± 1.68 | 0.0147 | 0.0697 | 27.2 | 30.1 | |
| miR-511 | 361.1 ± 1.35 | 2.6E-05 | 9.9E-04 | 40.0 | 31.6 | |
| miR-598 | −62.50 ± 1.22 | 8.0E-06 | 5.2E-04 | 33.9 | 40.0 | |
| p 24 h 0.5 Gy | miR-200a | −6,250.0 ± 1.38 | 1.1E-05 | 0.0020 | 27.4 | 40.0 |
| miR-292-3p | −137.0 ± 1.37 | 9.9E-05 | 0.0091 | 32.9 | 40.0 | |
| miR-379 | −123.5 ± 1.62 | 5.6E-04 | 0.0341 | 33.1 | 40.0 | |
| p 24 h 1.0 Gy | miR-342-5p | −1,790.0 ± 1.41 | 2.7E-05 | 0.0012 | 29.2 | 40.0 |
| miR-379 | −122.0 ± 1.63 | 6.1E-04 | 0.0226 | 33.1 | 40.0 | |
| miR-598 | 107.1 ± 1.19 | 1.1E-05 | 7.1E-04 | 40.0 | 33.2 | |
| miR-667 | 302.4 ± 1.06 | 1.0E-07 | 9.3E-06 | 40.0 | 31.7 | |
| miR-685 | 1,830.0 ± 1.09 | 1.0E-07 | 9.3E-06 | 40.0 | 29.1 | |
| miR-741 | 33.6 ± 1.63 | 0.0020 | 0.0617 | 40.0 | 34.9 |
ID, identification; SEM, standard error of the mean; FDR, false discovery rate.
Compared to the non-irradiated control sample.
Average CT values of non-irradiated control and irradiated samples, respectively.
Development of miRNA-based radiation class predictors
BRB-ArrayTools (Simon et al. 2007) was also used to perform class prediction using the ΔCT values of the differentially expressed miRNA. The nearest-centroid method was used as the prediction method. The centroid of each class is a vector containing the averages of the ΔCT values of the miRNA belonging to the class. The distance of the miRNA expression profile of a sample to each of the centroids is determined, and the sample is assigned to the class with the shortest distance. The robustness of the class predictor was tested by calculating the misclassification error rate using the leave-one-out cross-validation method. Both of our classifiers (radiation type and radiation dose) produced error P-values of less than 0.0001, with 10,000 random permutations.
Gene ontology analysis of miRNA target genes
miRNA target genes were determined using TargetScan-Mouse v. 5.2 (Lewis et al. 2005). Only phylogenetically conserved miRNA targets with a context score ≤ −0.3, corresponding to a log2 gene expression ratio of ≤ −0.3, as determined by multivariate linear regression fitting of gene expression microarray data (Grimson et al. 2007, Friedman et al. 2009), were used for gene-ontology (GO) analysis.
The predicted miRNA target genes were uploaded to the PANTHER GO database v. 7.0 (Thomas et al. 2003), which classifies genes according to GO terms, using published scientific experimental evidence. Gene set enrichment analysis was performed, with the NCBI mouse gene list serving as the reference whole-genome list. A binomial test was used for pathway enrichment analysis.
Results
miRNA expression signatures induced by irradiation
A total of 119 mouse miRNA (out of an average 188 per sample that were amplified above background level) were differentially expressed with P-values of less than 0.05. The differentially expressed miRNA were subjected to an FDR analysis, which controls for the expected proportion of false positives (Benjamini and Hochberg 1995). The FDR was set to 0.07 which means that only 7% of the miRNA declared as differentially expressed are expected to be false positives. After applying this restrictive adjustment, we determined that 26 miRNA were differentially expressed (FDR < 0.07, average P-value of 0.0002; Table I). The differentially expressed miRNA were expressed in either one condition (77%) or multiple conditions (23%). Notably, the majority of differentially expressed miRNA (79%) were downregulated after proton irradiation.
The results from the power analysis show that we have relatively low power to detect 2-fold differences between the compared experimental groups (geometric mean of 0.23). The power improved for differences of 3-fold and over and is 0.69 for 3-fold differences, 0.88 for 4-fold differences, and 0.95 for 5-fold differences. Accordingly, 24 out of the 26 detected differentially expressed miRNA exhibit a radiation-induced up- or downregulation of 3-fold or more.
Comparison of miRNA signatures for different types and energies of radiation
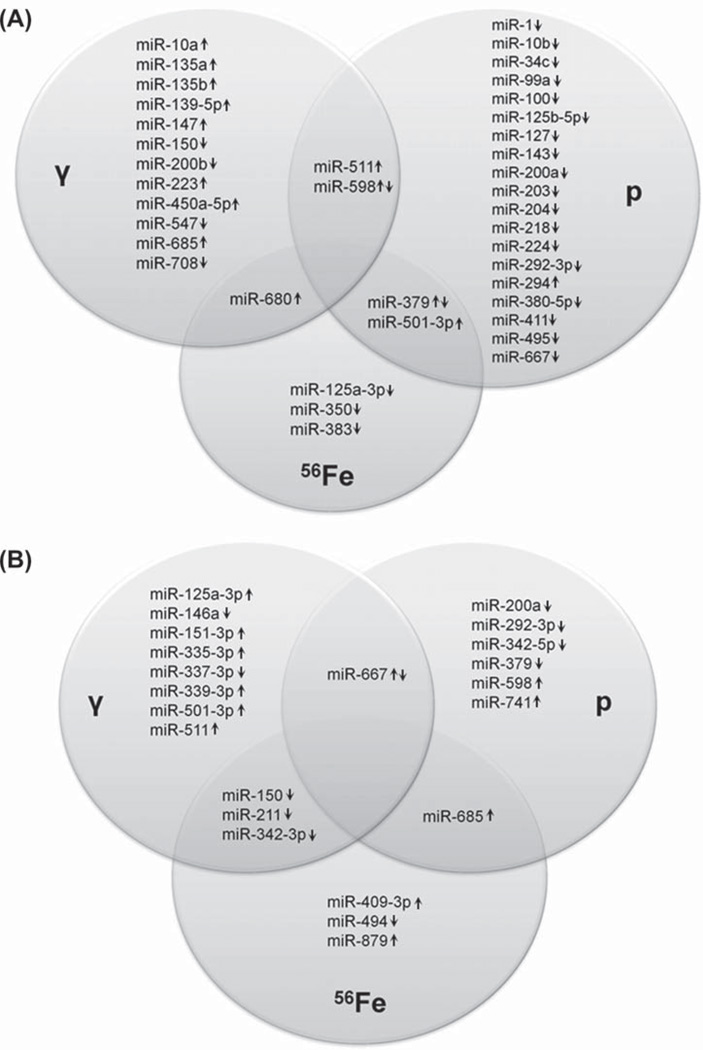
In a previous study using the same mouse strain (from the same vendor) and experimental conditions, we assessed the miRNA response to γ-rays and 1 GeV/n 56Fe ions. As compared to the 600 MeV proton irradiations, the prior 56Fe-ion particle exposures had comparable relative biological effectiveness (RBE) magnitudes. Data in the previous study was processed with the same criteria for statistical significance (Templin et al. 2011a). Based on these factors, we believe that a reliable comparison between the two studies is possible. A comparison of the differentially expressed miRNA shows that a total of 39 miRNA are differentially expressed 6 h after irradiation with γ-rays, protons, or 56Fe ions and that 22 miRNA are differentially expressed 24 h after exposure to any of these three radiation types. For each time point, 5 miRNA are differentially expressed upon exposure to either of two different types of radiation, amounting to, respectively, 12.8% and 22.7% of all differentially expressed miRNA at the 6 h and 24 h time point, but no miRNA are differentially expressed in response to all three radiation types (Figure 1).
Figure 1.
Venn diagrams showing miRNA differentially expressed upon irradiation with γ-rays, 600 MeV protons, or 1 GeV/n 56Fe ions, regardless of radiation dose, at 6 h (A) or 24 h (B) after radiation exposure. Areas of overlap among different circles depict miRNA differentially expressed after exposure to either of two different types of radiation. Arrows indicate up or down regulation of the specific miRNA after irradiation.
miRNA signature-based class prediction
The differentially expressed miRNA obtained in this study together with the miRNA expression results obtained in our previous study were used to build classifiers in order to test to what degree blood miRNA signature-based class prediction is able to indicate the irradiation status of mice. The leave-one-out cross validation method was used to test the robustness of the classifiers. In this method, all samples – except one – are used to build the classifier, and the ability of the classifier to correctly predict the class membership of the left-out sample is computed. This process is continued until each sample has been left out once, and the parameters that characterize the performance of the classifier are based on the overall ability of the classifier to correctly assign the samples to their respective classes.
Three parameters were calculated to gauge classifier performance: Accuracy, sensitivity, and specificity. Accuracy is the percentage of samples correctly assigned to the class they belong to. Sensitivity is the probability for a sample belonging to a class to be correctly predicted as belonging to that class. Specificity is the probability for a sample not belonging to a class to be correctly predicted as not belonging to that class.
Three different types of classifiers were created. The performance of these classifiers is illustrated in Table II. The first classifier was designed to classify samples according to radiation type (γ, proton, 56Fe, or control), irrespective of time point after irradiation and radiation dose. Thirty-two miRNA were included in this predictor. This predictor correctly classified 81% of the unknown samples, with specificities and sensitivities ranging from 0.583–1.00.
Table II.
Performance of class-prediction classifiers. Accuracy, sensitivity, and specificity of the class-prediction classifiers designed to predict the irradiation type, proton-irradiation dose or time after proton irradiation based on miRNA expression signatures.
| Irradiation condition | Accuracy | Sensitivity | Specificity |
|---|---|---|---|
| γ | 81% | 0.583 | 0.902 |
| p | 1 | 1 | |
| 56Fe | 0.75 | 0.902 | |
| Control | 0.882 | 0.944 | |
| p 0.0 Gy | 88% | 1 | 0.833 |
| 0.5 Gy | 1 | 1 | |
| 1.0 Gy | 0.667 | 1 | |
| p 6 h | 83% | 0.778 | 0.889 |
| p 24 h | 0.889 | 0.778 |
p = protons.
The second classifier was designed to classify samples according to the received proton radiation doses irrespective of the time point after radiation exposure. This type of predictor may be used for a sample known to be exposed to proton radiation. This predictor contained 3 miRNA (miR-292 - 3p, miR-379, and miR-667) and correctly classified 88% of the unknown samples, with specificities and sensitivities ranging from 0.667–1.00. The third classifier was designed to classify samples according to the time after receiving proton irradiation. It can be used for a sample known to be exposed to proton radiation and contained the same miRNA used in the dose prediction classifier. This predictor correctly classified 83% of the unknown samples with specificities and sensitivities ranging from 0.778–0.889.
It is important to consider that class predictors can be employed in combinations. For example, once the radiation type has been determined using this classifier, the miRNA expression data obtained from blood samples can be further classified using separate radiation type-specific predictors, such as the one for protons shown above or the γ ray- or 56Fe ion-specific predictors we developed previously (Templin et al. 2011a). We conclude that, in principle, blood miRNA expression profiles derived from C57BL/6 mice can be used to correctly predict the type and dose of radiation received by the animal.
miRNA targets and gene ontology analysis
The goal of the GO analysis was to determine the physiological significance of radiation-induced miRNA expression changes. This was a stepwise analysis, in which we initially determined the genes that are targeted by the differentially expressed miRNA, followed by GO analysis of the miRNA target genes. Radiation-induced miRNA expression changes in total blood are the result of two factors: (i) Changes in blood cell composition (because of cell killing) and (ii) cell-specific changes in miRNA expression. Therefore, a direct comparison of expression data from irradiated and non-irradiated blood samples is difficult because the cell composition in these two conditions is not the same, especially for high doses (Niemoeller et al. 2011, Nikiforova et al. 2011). To avoid a misinterpretation of expression data, we performed GO analysis of genes targeted only by those differentially expressed miRNA that (i) had no detectable expression in the control samples, and (ii) were switched on by radiation in the irradiated samples. The same strategy was used in our previous work.
miRNA target genes were determined using TargetScan-Mouse v. 5.2. Only high-probability target genes with context scores of ≤ − 0.3 and at least one conserved miRNA target site, as defined by phylogenetic-tree analysis, were selected. Using these criteria, only conserved target genes that were downregulated by at least 23% by the respective miRNA were included as bona fide targeted transcripts.
GO analysis for biological process and molecular function was performed using the PANTHER GO database v. 7.0 (Thomas et al. 2003). Since the PANTHER analysis platform had been updated, commensurate with the expansion of published interactions of signaling and effector pathways, we repeated the analysis of genes targeted by miRNA switched on by γ or 56Fe irradiation. The results suggest that several biological processes and molecular functions are modulated by the radiation-induced miRNA (Tables III and IV). Very importantly, the biological processes and molecular functions most significantly enriched in the predicted target genes of the switched-on miRNA were very similar among to the γ, 600 MeV proton-, and 56Fe-irradiation conditions. A survey of target genes suggests that up to 309 genes can be downregulated by the radiation-induced miRNA in the proton-irradiated group, 142 genes in the 56Fe ion-irradiated group, and 160 genes in the γ-irradiated group. Also, 130 target genes were common to both the proton- and γ-irradiated groups, while 52 genes overlapped in the proton- and 56Fe-irradiated groups. Finally, 80 genes overlapped in both γ- and 56Fe-irradiated samples. These findings highlight the similarities and differences in the cell response to different types of radiation and demonstrate that a characterization of radiation response biology can be obtained by miRNA target analysis.
Table III.
Biological processes putatively activated by γ-, 600 MeV proton, and 1 GeV 56Fe irradiation. We performed gene ontology analysis of genes targeted only by those differentially expressed miRNA that (1) had no detectable expression in the control samples, and (2) were switched on by radiation in the irradiated samples. Gene set enrichment analysis for biological processes was performed, using a binomial test. The first seven processes that showed the statistically most significant enrichment in the predicted target genes of the switched-on miRNA, as well as the test’s P-values, are shown.
| γ-rays | P-value | Protons | P-value | 56Fe ions | P-value |
|---|---|---|---|---|---|
| Nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | 5.10E-10 | Nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | 1.13E-26 | Primary metabolic process | 4.69E-12 |
| Metabolic process | 2.14E-09 | Primary metabolic process | 96E-18 | Metabolic process | 3.01E-11 |
| Primary metabolic process | 2.27E-09 | Metabolic process | 7.02E-16 | Nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | 4.45E-08 |
| Nervous system development | 1.37E-05 | Ectoderm development | 7.60E-08 | Cellular process | 7.41E-05 |
| Segment specification | 1.58E-05 | Nervous system development | 2.25E-07 | Apoptosis | 2.06E-04 |
| Ectoderm development | 2.69E-05 | System development | 2.48E-06 | Cell cycle | 3.94E-04 |
| Pattern specification process | 1.26E-04 | Cellular process | 1.18E-05 | Nervous system development | 4.99E-04 |
Table IV.
Molecular functions putatively activated by γ-, 600 MeV proton, and 1 GeV 56Fe irradiation. The first five functions that showed the statistically most significant enrichment in the predicted target genes of the switched-on miRNA, as well as the P-values of a binomial significance test, are shown.
| Molecular function | P-value γ | P-value p | P-value 56Fe |
|---|---|---|---|
| Nucleic acid binding | 1.22E-09 | 4.84E-24 | 7.65E-09 |
| DNA binding | 1.66E-08 | 1.07E-26 | 3.05E-09 |
| Binding | 3.21E-08 | 4.82E-21 | 8.73E-08 |
| Transcription factor activity | 1.40E-07 | 1.13E-24 | 1.68E-07 |
| Transcription regulator activity | 1.40E-07 | 1.13E-24 | 1.68E-07 |
p = protons.
Discussion
In this paper we analyzed miRNA expression in the blood of mice irradiated with 600 MeV protons. Our main finding is that proton irradiation induces changes in the miRNA signature of whole blood and that these changes depend on the irradiation parameters. Another finding is that miRNA expression profiles depend on radiation quality, i.e., every type of radiation induces its own distinctive miRNA signature. And lastly, the GO results suggest that the radiation-induced miRNA can be related through their target genes to similar GO categories. A practical outcome of our study is that it is possible to develop miRNA-based statistical classifiers. These classifiers exactly predict the radiation exposure of samples with previously unknown irradiation status, providing information on both radiation quality and dose. Overall, these results show that miRNA expression signatures can be used successfully in radiation biodosimetry.
The specificity of miRNA signatures for γ, high-LET proton and 56Fe irradiation, described in this study, is a new, but not surprising, finding. As shown over the last five years, miRNA signatures are disease-specific and have diagnostic and prognostic value frequently surpassing that of other methods. This is true for cancer (reviewed in Croce 2009, Ferracin et al. 2010) as well as many other diseases (reviewed in Hebert and De Strooper 2009, Latronico and Condorelli 2009, Nana-Sinkam et al. 2009, Kerr and Davidson 2010, Taft et al. 2010). Therefore, every cellular process that is maintained by gene expression may have specific miRNA expression signatures. Our findings provide more evidence that supports this concept.
We found that 26 miRNA (14% of all detected miRNA) were differentially expressed in mouse blood after 600 MeV proton irradiation. More miRNA were differentially expressed upon exposure to the higher dose of 1.0 Gy, compared to the lower dose of 0.5 Gy, and more miRNA were differentially expressed at the earlier time point of 6 h, compared to the later time point of 24 h. These findings may be explained by the extent of irradiation damage and concomitant repair. The differentially expressed miRNA fall into two categories: (1) miRNA expressed in only one condition, and (2) miRNA expressed in multiple conditions. Of the differentially expressed miRNA, 77% are specific to a particular radiation condition. The remainder is expressed in two conditions (19%) or three conditions (4%). Most miRNA are downregulated after proton exposure (79%), which may be explained by a combination of radiation-induced changes in blood-cell miRNA expression and cell killing. Furthermore, 5 miRNA that change in response to proton irradiation are also differentially expressed upon exposure to γ or 56Fe irradiation, at 6 or 24 h after radiation exposure. These findings illustrate the complexity of the radiation response and the importance of miRNA in this response. A comparison of the miRNA signatures obtained in this study and of the miRNA signatures induced by equitoxic doses of low-LET γ and high-LET 56Fe radiation (Templin et al. 2011a) shows that miRNA profiles are radiation type-specific. Consider that doses of 1.5 Gy γ, 1.0 Gy proton, and 0.5 Gy 56Fe irradiation have a similar RBE (Peng et al. 2009). All these irradiations induce different miRNA expression signatures. Remarkably, a subset of the differentially expressed miRNA becomes increased or decreased depending of the type of radiation used (Figure 1). Intriguingly, there is not a single miRNA that is modulated by all three types of radiation.
The results presented here are in agreement with previous studies that showed differences between gene expression signatures induced by low- and high-LET irradiation in cells and tissues (Ding et al. 2005, Zhang et al. 2006, Matsumoto et al. 2008). It should be noted that it would be useful for the use of miRNA in biological dosimetry to study the kinetics of miRNA regulation over a longer period than was done in our studies. Since we also examined the variables radiation type and radiation dose, we limited our studies to relative short time points after irradiation (6 and 24 h), in order to keep the number of investigated variables manageable. Cost constraints further limited the number of time points we were able to investigate. Future studies will hopefully address the radiation-induced miRNA response at longer time points after radiation exposure.
We attempted to cross-validate our results with other studies, but did not find any other published work describing radiation-induced miRNA expression changes in mouse blood for comparison. A comparison of our findings with radiation-induced miRNA expression profiles in human cell lines and primary cells shows very little overlap, probably because of differences in species, model system, cell type, and irradiation conditions (Maes et al. 2008, Cha et al. 2009a, 2009b).
Condition-specific gene expression signatures are frequently used to build statistical classifiers that can categorize unknown samples. Gene expression signatures of mouse and human peripheral blood cells were successfully used to build radiation class-prediction classifiers (Meadows et al. 2008, 2010, Paul and Amundson 2008). Based on the detected differences in miRNA expression signatures, we developed classifiers that can be used as predictors of exposure to γ-rays, protons or 56Fe ions. Our predictors classified the samples quite well according to radiation type (accuracy of 81%), proton dose (accuracy of 88%) and time after proton irradiation (accuracy of 83%). These classifiers encompass various scenarios that require predictions about received radiation exposures. An advantage is that the classifiers do not require a pre-exposure control sample because (i) all samples that were used to develop the classifiers were derived from separate, independent organisms, and (ii) the nearest-centroid classifiers that we developed are able to assign a radiation type and dose to a sample with unknown irradiation status based on the absolute normalized expression values (ΔCT values) of the miRNA contained in the classifier. It has to be noted, however, that the miRNA used for class prediction are not necessarily radiation-specific. Differential expression of miRNA as reported in this study might also be induced by other cellular stresses, such as inflammation, reactive oxygen species, and cytotoxic chemicals (Simone et al. 2009).
In addition to developing biodosimetric miRNA signatures, we identified biological processes and functions potentially impacted as a result of miRNA induction caused by radiation. The signaling networks that respond to this stimulus provide insights into the physiological changes in mouse blood induced by proton irradiation. The GO analysis results show that irradiation changes miRNA control of specific biological processes and molecular functions, such as DNA binding, transcription factor activity, metabolic processes, and others. There is a large overlap between the genes targeted in the γ-, proton-, and 56Fe ion-irradiated blood samples. Accordingly, a similar pattern of signal transduction developed as evidenced by the similarities in GO processes and functions. While the overlapping set of genes is not identical, there is sufficient commonality, which indicates a unified overall response to radiation. The evident differences among the target genes and GO categories, depending on irradiation type, point to a precise discrimination of the damage induced by the different types of radiation. Together, these results demonstrate a high level of resolution in the cellular response to radiation. Given that the determination of the miRNA target genes was carried out by software analysis, we risk overstating the significance of these findings. However, the rigorous and very conservative limits specified in our analyses support the biological validity of our findings. Future work in this area will include the validation of miRNA-mediated control of radiation response physiology.
Acknowledgements
We are grateful to the staff at the NASA Space Radiation Laboratory and at the Medical and Biology Departments of Brookhaven National Laboratory. In particular, we wish to thank Drs Adam Rusek, Michael Sivertz, and I-Hung Chiang for their help with dosimetry and irradiation of animals, and Dr Peter Guida for his organizational support. Additionally, we acknowledge the assistance of MaryAnn Petry and the staff of the Brookhaven Animal Facility. This study was supported by NASA, grant number NNX07AT41G, and by the Center for High-Throughput Minimally-Invasive Radiation Biodosimetry, National Institute of Allergy and Infectious Diseases, grant number U19 AI067773.
Footnotes
Declaration of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Aypar U, Morgan WF, Baulch JE. Radiation-induced epigenetic alterations after low and high LET irradiations. Mutation Research. 2011;707:24–33. doi: 10.1016/j.mrfmmm.2010.12.003. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society B. 1995;57:289–300. [Google Scholar]
- Cha HJ, Seong KM, Bae S, Jung JH, Kim CS, Yang KH, Jin YW, An S. Identification of specific microRNAs responding to low and high dose gamma-irradiation in the human lymphoblast line IM9. Oncology Reports. 2009a;22:863–868. [PubMed] [Google Scholar]
- Cha HJ, Shin S, Yoo H, Lee EM, Bae S, Yang KH, Lee SJ, Park IC, Jin YW, An S. Identification of ionizing radiation-responsive microRNAs in the IM9 human B lymphoblastic cell line. International Journal of Oncology. 2009b;34:1661–1668. doi: 10.3892/ijo_00000297. [DOI] [PubMed] [Google Scholar]
- Chaudhry MA. Real-time PCR analysis of micro-RNA expression in ionizing radiation-treated cells. Cancer Biotherapy & Radiopharmaceuticals. 2009;24:49–56. doi: 10.1089/cbr.2008.0513. [DOI] [PubMed] [Google Scholar]
- Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nature Reviews Genetics. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickey JS, Zemp FJ, Martin OA, Kovalchuk O. The role of miRNA in the direct and indirect effects of ionizing radiation. Nature Reviews Genetics. 2011;50:491–499. doi: 10.1007/s00411-011-0386-5. [DOI] [PubMed] [Google Scholar]
- Ding LH, Shingyoji M, Chen F, Chatterjee A, Kasai KE, Chen DJ. Gene expression changes in normal human skin fibroblasts induced by HZE-particle radiation. Radiation Research. 2005;164:523–526. doi: 10.1667/rr3350.1. [DOI] [PubMed] [Google Scholar]
- Ferracin M, Veronese A, Negrini M. Micromarkers: miRNAs in cancer diagnosis and prognosis. Expert Review of Molecular Diagnostics. 2010;10:297–308. doi: 10.1586/erm.10.11. [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Research. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Molecular Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert SS, De Strooper B. Alterations of the microRNA network cause neurodegenerative disease. Trends in Neuroscience. 2009;32:199–206. doi: 10.1016/j.tins.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Ishii H, Saito T. Radiation-induced response of micro RNA expression in murine embryonic stem cells. Medicinal Chemistry. 2006;2:555–563. doi: 10.2174/1573406410602060555. [DOI] [PubMed] [Google Scholar]
- Josson S, Sung SY, Lao K, Chung LW, Johnstone PA. Radiation modulation of microRNA in prostate cancer cell lines. Prostate. 2008;68:1599–1606. doi: 10.1002/pros.20827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr TA, Davidson NO. Therapeutic RNA manipulation in liver disease. Hepatology. 2010;51:1055–1061. doi: 10.1002/hep.23344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer A, Anastasov N, Angermeier M, Winkler K, Atkinson MJ, Moertl S. MicroRNA-mediated processes are essential for the cellular radiation response. Radiation Research. 2011;176:575–586. doi: 10.1667/rr2638.1. [DOI] [PubMed] [Google Scholar]
- Latronico MV, Condorelli G. MicroRNAs and cardiac pathology. Nature Reviews Cardiology. 2009;6:419–429. doi: 10.1038/nrcardio.2009.56. [DOI] [PubMed] [Google Scholar]
- Lee ML. Power and sample size considerations. Analysis of microarray gene expression data. Norwell, MA, USA: Kluwer Academic Publishers; 2004. pp. 193–234. [Google Scholar]
- Lee ML, Whitmore GA. Power and sample size for DNA microarray studies. Statistics in Medicine. 2002;21:3543–3570. doi: 10.1002/sim.1335. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Maes OC, An J, Sarojini H, Wu H, Wang E. Changes in MicroRNA expression patterns in human fibroblasts after low-LET radiation. Journal of Cell Biochemistry. 2008;105:824–834. doi: 10.1002/jcb.21878. [DOI] [PubMed] [Google Scholar]
- Marsit CJ, Eddy K, Kelsey KT. MicroRNA responses to cellular stress. Cancer Research. 2006;66:10843–10848. doi: 10.1158/0008-5472.CAN-06-1894. [DOI] [PubMed] [Google Scholar]
- Matsumoto Y, Iwakawa M, Furusawa Y, Ishikawa K, Aoki M, Imadome K, Matsumoto I, Tsujii H, Ando K, Imai T. Gene expression analysis in human malignant melanoma cell lines exposed to carbon beams. International Journal of Radiation Biology. 2008;84:299–314. doi: 10.1080/09553000801953334. [DOI] [PubMed] [Google Scholar]
- Meadows SK, Dressman HK, Muramoto GG, Himburg H, Salter A, Wei Z, Ginsburg GS, Chao NJ, Nevins JR, Chute JP. Gene expression signatures of radiation response are specific durable and accurate in mice and humans. PLoS One. 2008;3:e1912. doi: 10.1371/journal.pone.0001912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows SK, Dressman HK, Daher P, Himburg H, Russell JL, Doan P, Chao NJ, Lucas J, Nevins JR, Chute JP. Diagnosis of partial body radiation exposure in mice using peripheral blood gene expression profiles. PLoS One. 2010;5:e11535. doi: 10.1371/journal.pone.0011535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nana-Sinkam SP, Hunter MG, Nuovo GJ, Schmittgen TD, Gelinas R, Galas D, Marsh CB. Integrating the MicroRNome into the study of lung disease. American Journal of Respiratory and Critical Care Medicine. 2009;179:4–10. doi: 10.1164/rccm.200807-1042PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemoeller OM, Niyazi M, Corradini S, Zehentmayr F, Li M, Lauber K, Belka C. MicroRNA expression profiles in human cancer cells after ionizing radiation. Radiation Oncology. 2011;6:29. doi: 10.1186/1748-717X-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforova MN, Gandi M, Kelly L, Nikiforov YE. MicroRNA dysregulation in human thyroid cells following exposure to ionizing radiation. Thyroid. 2011;21:261–266. doi: 10.1089/thy.2010.0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Amundson SA. Development of gene expression signatures for practical radiation biodosimetry. International Journal of Radiation, Oncology Biology Physics. 2008;71:1236–1244. doi: 10.1016/j.ijrobp.2008.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Brown N, Finnon R, Warner CL, Liu X, Genik PC, Callan MA, Ray FA, Borak TB, Badie C, et al. Radiation leukemogenesis in mice: Loss of PU.1 on chromosome 2 in CBA and C57BL/6 mice after irradiation with 1 GeV/nucleon 56Fe ions, X rays or gamma rays. Part. I. Experimental observations. Radiation Research. 2009;171:474–483. doi: 10.1667/RR1547.1. [DOI] [PubMed] [Google Scholar]
- Pradervand S, Weber J, Lemoine F, Consales F, Paillusson A, Dupasquier M, Thomas J, Richter H, Kaessmann H, Beaudoing E, et al. Concordance among digital gene expression, microarrays, and qPCR when measuring differential expression of microRNAs. Biotechniques. 2010;48:219–222. doi: 10.2144/000113367. [DOI] [PubMed] [Google Scholar]
- Shin S, Cha HJ, Lee EM, Lee SJ, Seo SK, Jin HO, Park IC, Jin YW, An S. Alteration of miRNA profiles by ionizing radiation in A549 human non-small cell lung cancer cells. International Journal of Oncology. 2009;35:81–86. [PubMed] [Google Scholar]
- Shkumatava A, Stark A, Sive H, Bartel DP. Coherent but overlapping expression of microRNAs and their targets during vertebrate development. Genes Development. 2009;23:466–481. doi: 10.1101/gad.1745709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R, Lam A, Li MC, Ngan M, Menenzes S, Zhao Y. Analysis of gene expression data using BRB-array tools. Cancer Information. 2007;3:11–17. [PMC free article] [PubMed] [Google Scholar]
- Simone NL, Soule BP, Ly D, Saleh AD, Savage JE, Degraff W, Cook J, Harris CC, Gius D, Mitchell JB. Ionizing radiation-induced oxidative stress alters miRNA expression. PLoS One. 2009;4:e6377. doi: 10.1371/journal.pone.0006377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: Regulators of disease. Journal of Pathology. 2010;220:126–139. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- Templin T, Amundson SA, Brenner DJ, Smilenov LB. Whole mouse blood microRNA as biomarkers for exposure to -rays and (56) Fe ion. International Journal of Radiation Biology. 2011a;87:653–662. doi: 10.3109/09553002.2010.549537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templin T, Paul S, Amundson SA, Young EF, Barker CA, Wolden SL, Smilenov LB. Radiation-induced micro-RNA expression changes in peripheral blood cells of radiotherapy patients. International Journal of Radiation Oncology, Biology, Physics. 2011b;80:549–557. doi: 10.1016/j.ijrobp.2010.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. PANTHER: A library of protein families and subfamilies indexed by function. Genome Research. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu W, Ting Lee M-L, Whitmore GA. Sample Size and Power Calculation in Microarray Studies Using the sizepower package. 2012 http://www.bioconductor.org/packages/devel/bioc/html/sizepower.html. [Google Scholar]
- Wright GW, Simon RM. A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics. 2003;19:2448–2455. doi: 10.1093/bioinformatics/btg345. [DOI] [PubMed] [Google Scholar]
- Yauk CL, Rowan-Carroll A, Stead JD, Williams A. Cross-platform analysis of global microRNA expression technologies. BMC Genomics. 2010;11:330. doi: 10.1186/1471-2164-11-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Burns FJ, Chen H, Chen S, Wu F. Alterations in gene expression in rat skin exposed to 56Fe ions and dietary vitamin A acetate. Radiation Research. 2006;165:570–581. doi: 10.1667/RR3556.1. [DOI] [PubMed] [Google Scholar]