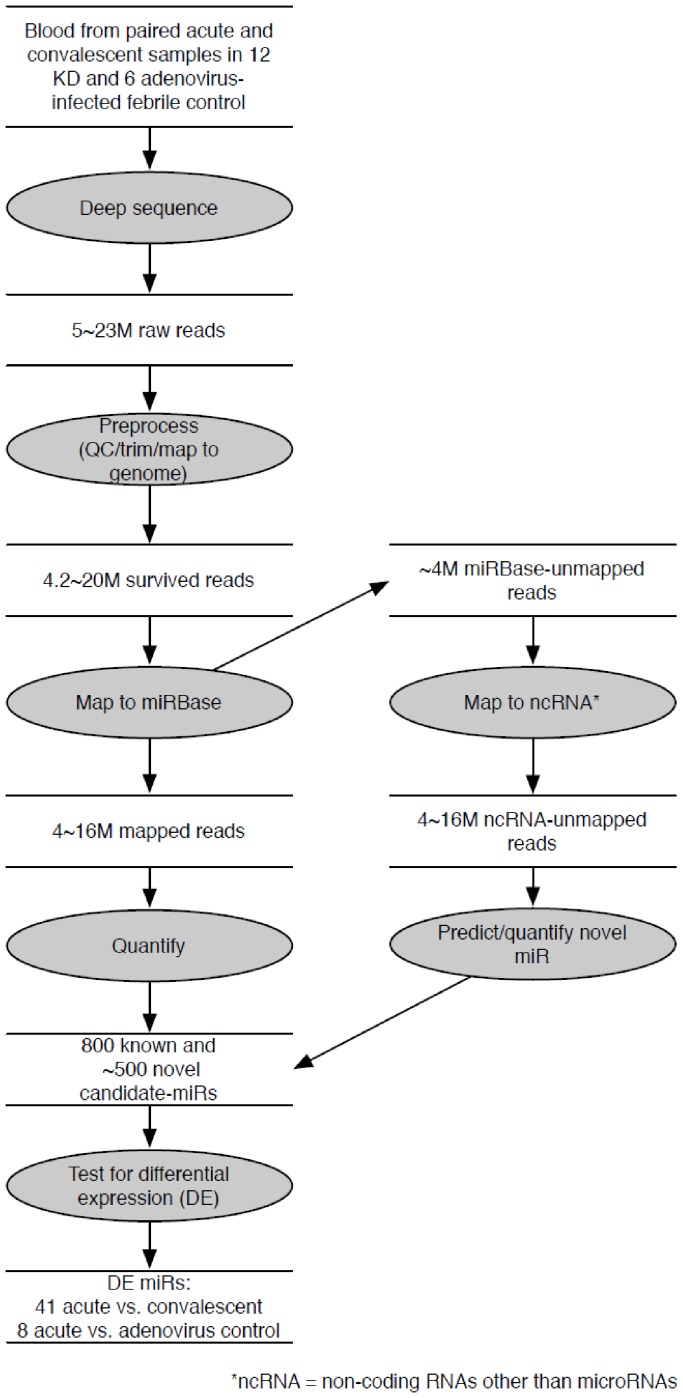
Figure 1. Flowchart of analytic strategy used to annotate known miRs and find differentially expressed miRs.
Total RNA extracted from whole blood from subjects with KD and acute adenovirus infection was sequenced using Illumina HighSeq. A custom-designed in-house tool identified known and novel miRs from among the small non-coding RNA species and analyzed differential expression of known miRs between acute vs. convalescent KD samples and acute KD vs. adenovirus-infected samples.