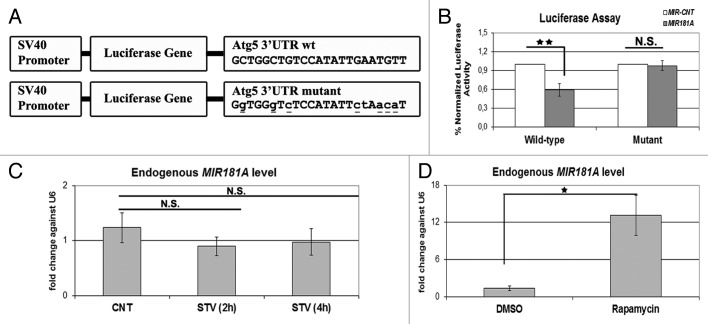
Figure 6. ATG5 as a direct target of MIR181A and changes in endogenous MIR181A levels during cellular stress. (A) A scheme representing luciferase constructs with wild-type (wt) or mutant 3′ UTR MIR181A MRE sequences of ATG5. Mutations are marked in lower case letters and underlined. (B) Normalized luciferase activity in lysates from 293T cells co-transfected with wild-type or mutant ATG5-luciferase constructs and MIR181A or MIR-CNT (mean ± SD of independent experiments, n = 3, **p < 0.01. N.S., not significant). (C) TaqMan quantitative PCR (qPCR) analysis of endogenous MIR181A levels under control (CNT, no starvation) or starvation (STV, 2 h and 4 h) conditions. Endogenous MIR181A levels were not responsive to starvation treatment (mean ± SD of independent experiments, n = 3, N.S., not significant). TaqMan qPCR data were normalized using U6 small nuclear 1 (RNU6-1) (U6) mRNA levels. (D) TaqMan qPCR analysis of endogenous MIR181A levels in cells treated with DMSO (carrier) or rapamycin. Endogenous MIR181A levels were increased following rapamycin treatment (mean ± SD of independent experiments, n = 4, *p < 0.05).

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.