Abstract
Human arginase is a binuclear manganese metalloenzyme that participates in the urea cycle. Arginase catalyses the hydrolysis of L-arginine into L-ornithine and urea and is linked to several disorders such as asthma and cancer. Currently, the protonation and tautomerization state of the substrate when bound to the active site, which contains two manganese ions, is not known. Knowledge of the charge dependent behavior of arginine in the arginase I environment would be of utility towards understanding the catalytic mechanism and designing inhibitors of this enzyme. The arginine+/0 species, including all possible neutral tautomers, were modeled using an aminoimidazole analog as template. All-atom molecular dynamics simulations were then performed on each of the charged and neutral species. In addition, a hydroxide ion was included in selected simulations to test its importance. Results show that the positively charged state of arginine is stable in the active site of arginase I, with that stabilization facilitated by the presence of hydroxide. Glu277 is indicated to play a role in stabilizing arginine in the active site and facilitating its ability to assume a catalytically competent conformation in the presence of hydroxide. The reported interactions and modeled arginine bound arginase I structures can be used as a tool for structure based inhibitor design as experimental data on the structure of the substrate-enzyme complex is lacking.
Keywords: manganese, arginine, Molecular Dynamics, molecular modeling, CHARMM, hydroxide ion
Introduction
Arginase is a trimeric binuclear manganese metalloenzyme that hydrolyzes L-arginine to L-ornithine and urea.1–3 Arginase has two iosforms that have 55% sequence identity and differ in subcellular localization and tissue distribution.2 Arginase I is located in the liver and red blood cells and catalyzes the final step of the urea cycle. Arginase II is involved in NO synthesis4 and distributed in the kidney, brain, gastrointestinal tract and prostate tissues.1 Arginase I plays a critical role in modulating the immune response,5, 6 and its inhibition can block lung carcinoma7, whereas arginase II is primarily involved in L-arginine homeostasis.8 Depletion of L-arginine by increased arginase activity causes allergic asthma.9, 10 L-arginine is also a substrate for nitric oxide synthase (NOS) where NO is produced when L-arginine is converted to L-citrulline by NOS.11
Arginase inhibitors can increase the concentration of L-arginine, enhancing NO biosynthesis and NO-dependent physiological processes such as smooth muscle relaxation. For example, long term treatment of spontaneously hypertensive rats with the arginase inhibitor Nω-hydroxy-L-nor-arginine results in reduced blood pressure, improved vascular function, and reduced cardiac fibrosis.12 In addition, NO regulates the nervous and immune systems13 and arginase inhibition is therapeutically beneficial in treating sexual arousal disorders.2 Additionally, inhibition of malarial arginase is beneficial in antimalarial therapy,14 the inhibitor Nω-hydroxy-L-arginine has been reported to inhibit the infection of the Leishmania parasite,15 and another inhibitor, L-norvaline, prevents endothelial dysfunction.16 Accordingly, inhibition of arginase has a range of potential therapeutic applications.
The active site of arginase contains two Mn2+ ions (Mn2+A – Mn2+B) separated by 3.3 Å, which are coordinated by Asp and His residues.17 These ions facilitate formation of a hydroxide ion that is essential in the enzyme’s catalytic mechanism. Replacement of the metal coordinating residues in arginase I disrupts the metal cluster and/or the nucleophilic hydroxide ion.18 Mn2+B and Asp-128 alone can hold the hydroxide ion in a position to attack the scissile guanidium carbon. However, the additional Mn2+ increases the polarizability of the hydroxide ion, thereby facilitating its reactivity.
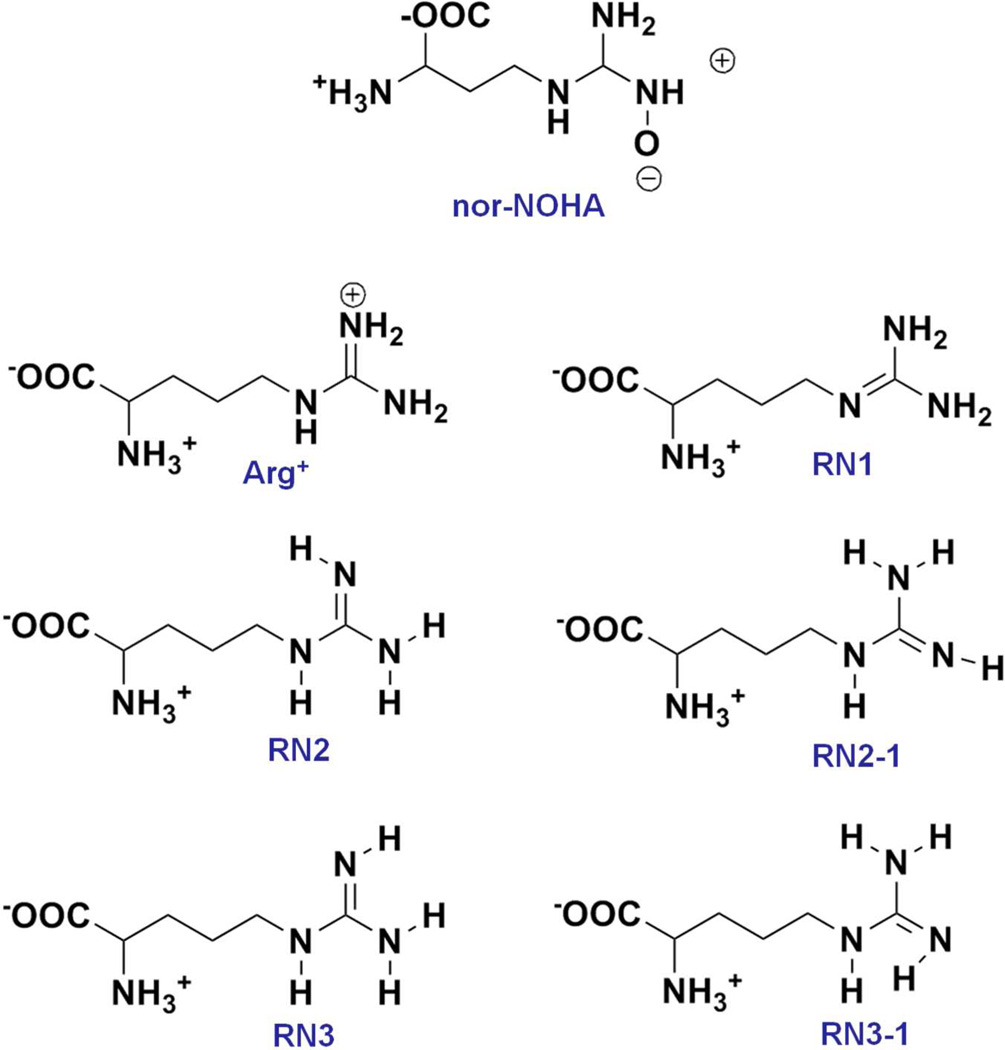
Molecular details of the interaction of various arginase inhibitors have been obtained from crystallographic studies.15, 19 For example, aminoimidazole based inhibitors and Nω-hydroxy-L-nor-arginine (nor-NOHA) complexes contain functional groups that can have direct metal coordination with the two Mn2+ ions in the catalytic site.19, 20 However, these structures do not yield information on how the substrate arginine binds in the active site. This includes consideration of the protonation state of the guanidinium group of arginine, which despite being in the pKa range of 12 – 13.7,21–23 could assume a neutral state when in the environment of the active site. And if a neutral state was assumed the arginine side chain can assume three possible tautomeric forms (Figure 1). In addition, given the role of the hydroxide ion, and the inability of available X-ray crystal structures to differentiate hydroxide from water, information on its role in substrate hydrolysis is lacking. In order to understand the binding mode and key interactions of substrates/inhibitors with arginase, including information on the presence of the nucleophilic hydroxide ion, we present a molecular dynamics (MD) simulation study of arginine and a selected inhibitor in the arginase I active site, with hydroxide included in a subset of the simulations. Results from the simulations show the role of a hydroxide ion in maintaining the proper distance between the metal ions, as well as maintain a functional metal cluster including the orientation of surrounding residues in the active site that is critical for hydrolysis.
Figure 1.
Inhibitor and substrate protonation and tautomeric states subjected to molecular dynamics simulation.
Materials and Methods
Computations were performed with version 35 of the CHARMM MD package24 with production simulations carried out using NAMD 2.8.25 Calculations were performed using the CHARMM22/CMAP26 all-atom additive force field supplemented with the CHARMM General Force Field27 for the inhibitor. Previously published ARG0 parameters28, 29 were used to treat the five possible tautomers of neutral arginine; RN1, RN2, RN-2, RN3 and RN3-1. These are pictorially represented in Figure 1 along with the studied inhibitor Nω-hydroxy- L-nor- arginine (nor-NOHA).20 The substrate arginine was modeled into the protein based on the aminohistidine inhibitor-arginase I crystal structure (PDB: 3MFV).19 The inhibitor ((2S)-2-amino-4-(2-amino-1H-imidazol-5-yl)butanoic acid) contains a 2-aminoimidazole heterocycle instead of a guanidinium group, such that deleting carbon 5 in the imidazole ring yields arginine (see Supplementary Figure S1) following adjustment of the geometry to remove the internal steric clash using the Clean function in Discovery Studio (Accelrys Inc.). The Arg+/0 species substrates were simulated with and without hydroxide ion (Figure 2). When hydroxide was included in the simulations it was initially positioned to symmetrically bridge both metal ions as observed for an inhibitor oxygen atom in the 2(S)-amino-6-boronohexanoic acid (ABH) bound crystal structure (PDB: 2AEB).30 For the metal ions, Mg2+ ion parameters were used to model the Mn2+ ions. To test the use of Mg2+ parameters a simulation was performed on the nor-NOHA-Arginases I complex; nor-NOHA is a competitive inhibitor with Kd = 51 nM (ITC) crystallized at a resolution of 1.66 Å (3KV2).20 The Nζ-OH group of nor-NOHA interacts symmetrically with metal ions, replacing the metal bridging hydroxide ion. Additionally, nor-NOHA interacts with protein by both direct and water mediated hydrogen bonds.
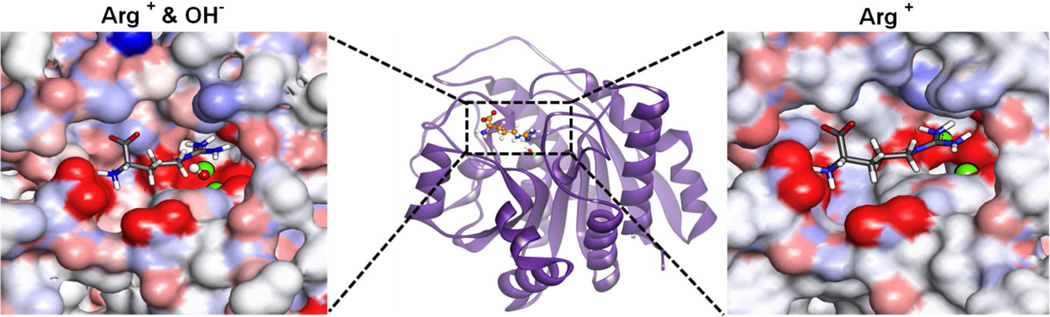
Figure 2.
Representative structures of Arginine (stick figure) bound to Arginase I with (left panel) and without (right panel) hydroxide ion (atom colored CPK). The Mg2+ ions are shown as green vdW spheres. Images created using Discovery Studio 3.0 (Accelrys Inc). Structures were obtained following modeling of substrate arginine based on the aminohistidine analogue inhibitor Z70 (PDB: 3MFV).
Protein preparation involved trimming the C-terminal 13 residues as their presence significantly increased the system size and they are ~25 Å away from the active and, therefore, not involved in substrate binding. The histidine protonation states were assigned based on Reduce.31 Generation of CHARMM inputs for the simulations, including building of hydrogens, system solvation and periodic boundary definition was carried using the CHARMM-GUI.32 The substrate/inhibitor-Arginase complexes were solvated with TIP3P water molecules33 in a cubic box extending a minimum of 10 Å beyond the protein non-hydrogen atoms and chloride ions were added to neutralize the systems (Table S1).
Simulation conditions included nonbonded interactions truncated using the force switching function between 8.0 and 10.0 Å for the Lennard-Jones interactions, and nonbonded list generation was restricted to 12.0 Å and was updated heuristically. Long range electrostatics were treated with the Particle Mesh Ewald (PME) method with the order set to 6 and kappa = 0.45 Å−1. The SHAKE method was applied to constrain the covalent bonds involving hydrogen atoms with an integration time step of 2 fs. All systems were minimized by 500 steps using the steepest descent minimizer with protein non-hydrogen atoms restrained by applying harmonic restraints of 5.0 kcal/mol/Å. Following that a 500 ps MD equilibration was carried in the NVT ensemble at 300 K with the non-hydrogen atoms restrained as described above. The equilibration allows for relaxation of the solvent and newly added hydrogens on the protein. Following the equilibration a 100 steps minimization was carried out with adopted basis Newton-Raphson minimizer without any restraints on the protein, inhibitors or solvent. The production simulations were performed in the NPT ensemble, temperature and pressure was maintained by the Langevin Piston method at 300 K and 1 atm, respectively. Initial atom velocities were assigned according to the Maxwell distribution. The initial 2 ns of each trajectory was considered additional equilibration and snapshots were saved every 5 ps from the remaining 18 or 48 ns production trajectories for analysis.
The trajectories obtained with the NAMD program were analyzed within the CHARMM program. Hydrogen bonds were defined as being present based on non-hydrogen (ie. O-N, O-O, or N-N) distance of 3.4 Å. VMD34 was used to visualize the trajectories obtained from MD simulations. For statistical analysis the 48 ns production trajectories were split into 4 blocks and 18 ns simulation into 3 and the block averages and standard errors calculated.
Results and Discussion
In total, thirteen MD simulations were conducted. The positive and neutral forms of the Arg guanidinium sidechain (ie. Arg+/0) were modeled based on the aminoimidazole-arginase complex structure (PDB: 3MFV)19 and MD simulations were carried for 20 or 50 ns of which the initial 2 ns were considered equilibration and the final 18 or 48 ns used for analysis. The role of the hydroxide ion was tested by performing each Arg+/0 simulation in the presence and absence of hydroxide (Figure 2). The active-site cleft is characterized by a Mn2+ - Mn2+ cluster that is essential for the catalytic activity of the arginase. As force field parameters for manganese ion were lacking, Mg2+ parameters were used due to its similar coordination preference. Initial analysis (see below) focused on the nor-NOHA-arginase simulation to verify the ability of the present simulation protocol to model the complex environment of the Arginase I active site. This was followed by analysis of the substrate Arg simulations in the different protonation and tautomeric states and in the presence and absence of hydroxide ion to understand the molecular nature of the substrate-protein interactions.
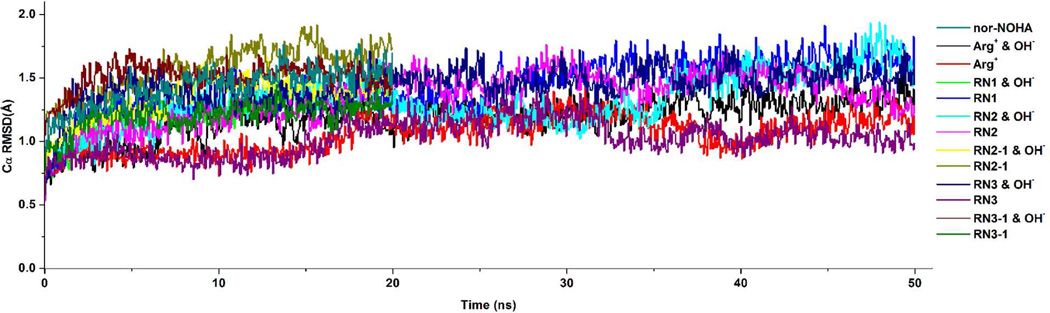
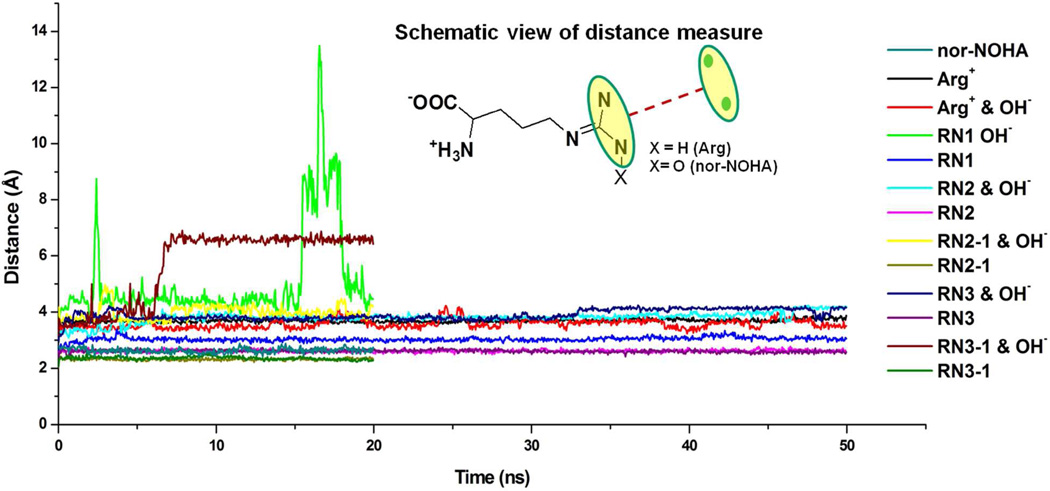
Convergence of all the simulation systems was analyzed by calculating the RMSD of the Cα atom of the protein with respect to the initial structures (Figure 3). The overall RMSD analysis indicates that the simulation systems were adequately converged after 2 ns, with final RMSD values below 2.0 Å. Variations in the RMSD as a function of simulation time are mainly due to the fluctuation of several loops connecting secondary elements in the protein as can been seen from the RMS fluctuations over the simulation as a function of residue for the nor-NOHA simulation (Figure S2, supporting information). The lowest RMSD values were observed for the Arg+-OH− complex. It should also be noted that in the RN1, RN2-1 and RN3-1 neutral substrate simulations with hydroxide, analysis of the guanidinium-Mg2+ distances (see below) indicates that the substrate is starting to diffuse from the binding site. However, this was not evidenced in the overall RMSD analysis.
Figure 3.
RMS deviation of the Cα atoms of the protein for the simulations conducted with the substrates and inhibitor nor-NOHA. RMSD calculation excluded the flexible C terminal loop 302 to 309 of the protein as it 19 Å or more from the active site metals ions.
Metal cluster structure in the presence of nor-NOHA
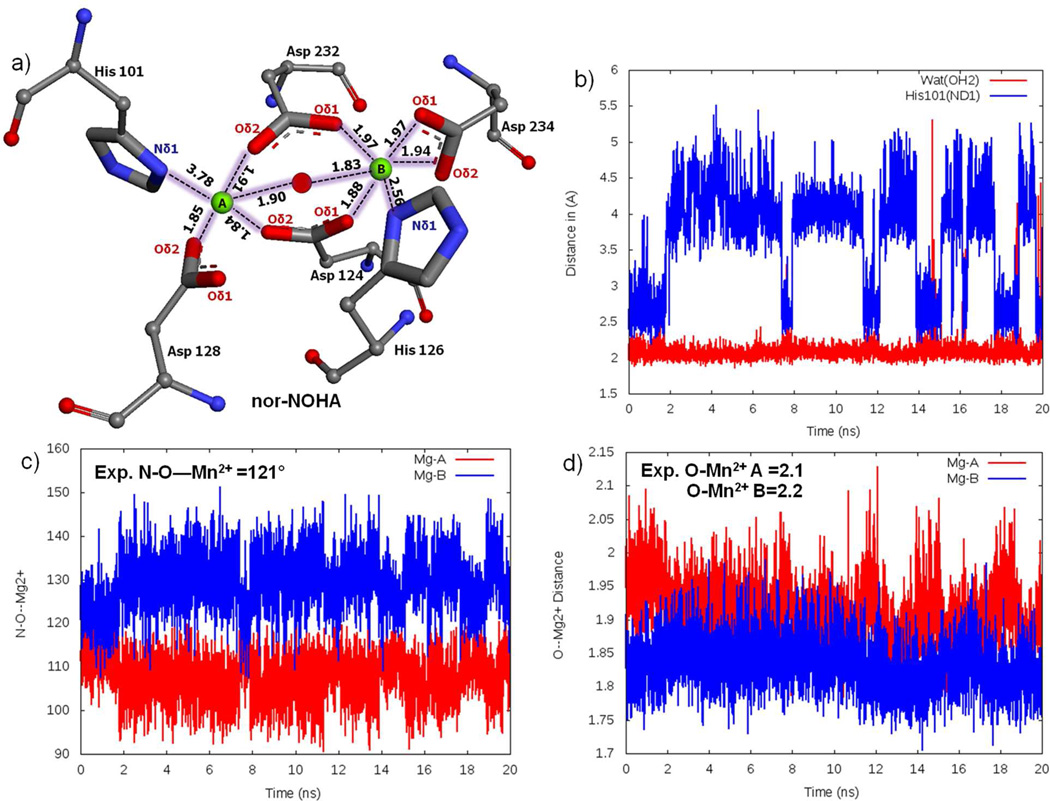
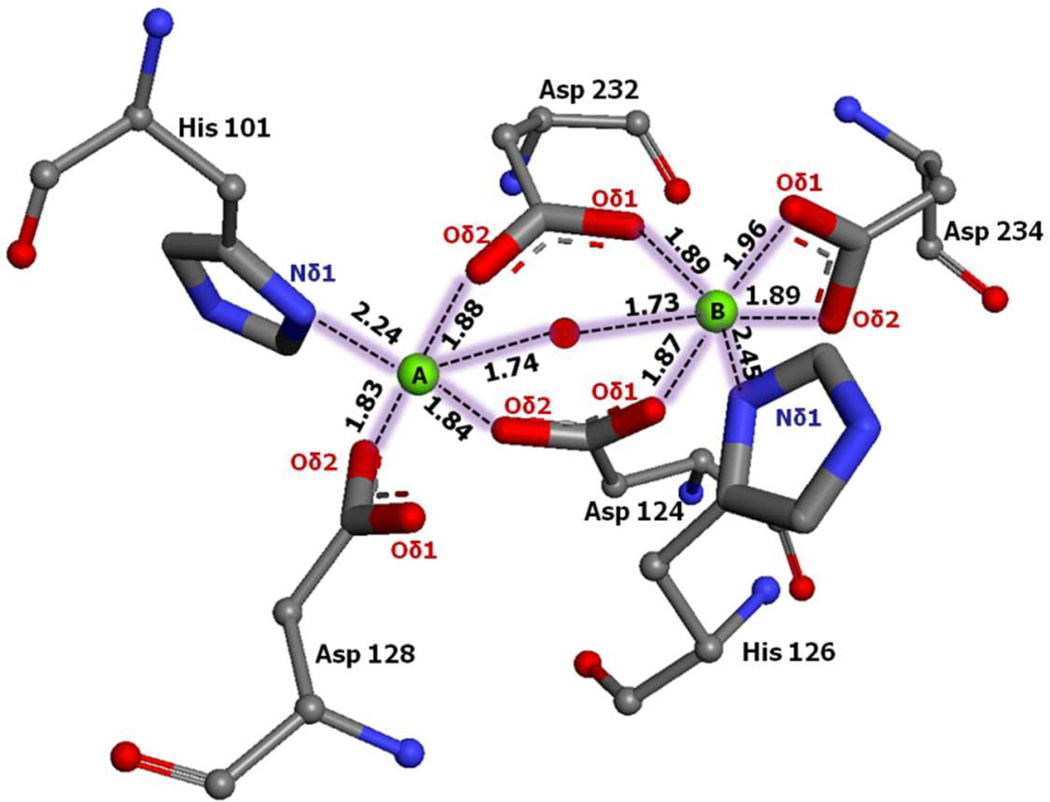
To demonstrate the reliability of the simulation protocol, including the use of Mg2+ parameters for Mn2+, the nor-NOHA simulation was analyzed and compared with the experimental data20. Analysis mainly focused on distances involving the metal ions, including hydrogen bonds established with protein atoms and water molecules. MgA has been observed to form a square pyramidal coordination with His101 (ND1), Asp124 (OD2), Asp128 (OD2), Asp232 (OD2) and Nω-hydroxy oxygen; the average distance for these atoms are 3.78±0.25 Å, 1.84±0.01 Å, 1.85±0.02 Å, 1.91±0.00 Å and 1.90±0.01 Å, respectively (Figure 4a and Table 1). Metal ion MgB is coordinated in an octahedral geometry with His126 (ND1), Asp124 (OD1), Asp234 (OD1 and OD2 bidentate), Asp232 (OD1) and N-(ω)-hydroxy oxygen, the distances obtained from the simulations between metal and ligand atoms are 2.56±0.04 Å, 1.88±0.02 Å, 1.97±0.02 Å, 1.94±0.01 Å and 1.83±0.01 Å, respectively. The simulated distances are generally comparable with the experimental values. An exception is the MgA-His101 average distance of 3.78 Å, which is longer than the 2.3 Å experimental distance. The reason for the His101 shift is competition of a water molecule with MgA that causes a shift in the location of the imidazole side chain. The nature of this competition is shown in Figure 4b, where towards the end of the simulation there are times when His101 moves close to the ion while the water moves away. Also, in the simulation a shift in the position of Asp 232 occurred. In the experimental structure Asp 232 Oδ2 symmetrically bridges the two metal ions, being 2.3 Å from each ion, whereas, in the simulation Oδ2 interacts primarily with MgA with a distance of 1.91±0.00 Å while Oδ1 shifts closer to MgB, forming an interaction with a distance of 1.97±0.02 Å. Additionally, the average coordination angles form the simulation for MgA (107±0.66°) and MgB (130±0.56°) are in satisfactory agreement with the experimentally determined coordination angle of N-O-Mn2+ 121° (Figure 4c). Similarly, the O-Mg2+ coordination distance is comparable with experimental O-Mn2+ distance (Figure 4d and Table 2). The above analysis indicates that the metal ions and surrounding protein maintain the experimentally determined geometry of the active site without applying any additional restraints to maintain the geometry.
Figure 4.
a) Magnesium cluster distances from the nor-NOHA simulation, Mg+2 ions A and B are shown as green spheres and the oxygen of the hydroxide is shown as a red sphere. b) Interference of water molecule in altering His 101 coordination. c) Coordination angle measure of N-O-Mg atoms. d) Distance between metal ions and metal bridging oxygen
Table 1.
Distance between the atoms coordinating the two Mg2+ ions.
| His101 Nδ1- MgA |
Asp128 Oδ1- MgA |
Asp128 Oδ2- MgA |
Asp124 Oδ1- MgA |
Asp124 Oδ2- MgA |
Asp232 Oδ1- MgA |
Asp232 Oδ2- MgA |
OH− - MgA |
|
|---|---|---|---|---|---|---|---|---|
| Experimental values | ||||||||
| Nor-NOHA | 2.3 | 3.6 | 2.2 | 3.5 | 2.1 | 4.4 | 2.3 | 2.0 |
| Simulation values | ||||||||
| Nor-NOHA | 3.78 ± 0.25 | 3.79 ± 0.08 | 1.85 ± 0.01 | 3.17 ± 0.04 | 1.84 ± 0.01 | 3.27 ± 0.02 | 1.91 ± 0.01 | |
| Arg+ & OH− | 2.24 ± 0.01 | 3.85 ± 0.01 | 1.83 ± 0.00 | 3.08 ± 0.01 | 1.84 ± 0.00 | 3.23 ± 0.01 | 1.88 ± 0.00 | 1.74 ± 0.00 |
| Arg+ | 2.02 ± 0.02 | 2.84 ± 0.41 | 1.81 ± 0.02 | 3.58 ± 0.03 | 1.80 ± 0.01 | 3.56 ± 0.02 | 1.79 ± 0.01 | |
| RN1 & OH− * | 4.89 ± 0.04 | 1.88 ± 0.00 | 2.00 ± 0.01 | 3.24 ± 0.01 | 1.87 ± 0.00 | 4.97 ± 0.12 | 3.39 ± 0.01 | 1.74 ± 0.00 |
| RN1 | 2.09 ± 0.00 | 1.89 ± 0.00 | 1.89 ± 0.00 | 3.90 ± 0.00 | 1.81 ± 0.00 | 6.13 ± 0.02 | 5.10 ± 0.08 | |
| RN2 & OH− | 4.54 ± 0.16 | 2.81 ± 0.30 | 1.85 ± 0.01 | 3.22 ± 0.02 | 1.91 ± 0.01 | 1.88 ± 0.01 | 3.17 ± 0.04 | 1.74 ±0.01 |
| RN2 | 2.07 ± 0.00 | 1.87 ± 0.01 | 1.87 ± 0.00 | 3.70 ± 0.01 | 1.80 ± 0.00 | 5.10 ± 0.02 | 3.68 ± 0.03 | |
| RN2-1 & OH− * | 2.28 ± 0.03 | 3.81 ± 0.01 | 1.82 ± 0.00 | 3.05 ± 0.02 | 1.86 ± 0.01 | 1.87 ± 0.01 | 3.23 ± 0.02 | 1.79 ± 0.01 |
| RN2-1* | 2.14 ± 0.01 | 3.70 ± 0.02 | 1.76 ± 0.01 | 3.61 ± 0.03 | 1.83 ± 0.03 | 3.74 ± 0.03 | 1.81 ± 0.01 | |
| RN3 & OH− | 4.64 ± 0.08 | 3.53 ± 0.22 | 1.84 ± 0.01 | 3.21 ± 0.02 | 1.91 ± 0.00 | 3.19 ± 0.05 | 1.90 ± 0.04 | 1.74 ± 0.01 |
| RN3 | 2.07 ± 0.00 | 1.86 ± 0.01 | 1.87 ± 0.01 | 3.71 ± 0.00 | 1.80 ± 0.00 | 5.08 ± 0.02 | 3.65 ± 0.01 | |
| RN3-1 & OH− * | 2.45 ± 0.07 | 3.80 ± 0.01 | 1.83 ± 0.01 | 3.14 ± 0.03 | 1.91 ± 0.04 | 3.22 ± 0.01 | 1.87 ± 0.01 | 1.80 ± 0.01 |
| RN3-1* | 2.12 ± 0.01 | 3.75 ± 0.02 | 1.76 ± 0.00 | 3.48 ± 0.03 | 1.77 ± 0.00 | 3.67 ± 0.04 | 1.82 ± 0.01 | |
| His126 Nδ1- MgB |
Asp234 Oδ1- MgB |
Asp234 Oδ2- MgB |
Asp232 Oδ1- MgB |
Asp232 Oδ2- MgB |
Asp124 Oδ1- MgB |
Asp124 Oδ2- MgB |
OH− - MgB |
|
| Experimental values | ||||||||
| Nor-NOHA | 2.3 | 2.3 | 2.3 | 3.7 | 2.3 | 2.1 | 3.3 | 2.3 |
| Simulation values | ||||||||
| Nor-NOHA | 2.56 ± 0.03 | 1.97 ± 0.02 | 1.94 ± 0.01 | 1.97 ± 0.02 | 3.37 ± 0.00 | 1.88 ± 0.02 | 3.39 ± 0.07 | |
| Arg+ & OH− | 2.45 ± 0.02 | 1.96 ± 0.00 | 1.89 ± 0.00 | 1.89 ± 0.00 | 3.10 ± 0.01 | 1.87 ± 0.00 | 3.24 ± 0.01 | 1.73 ± 0.00 |
| Arg+ | 2.06 ± 0.01 | 1.91 ± 0.00 | 1.85 ± 0.01 | 1.79 ± 0.00 | 3.39 ± 0.01 | 1.82 ± 0.00 | 3.47 ± 0.03 | |
| RN1 & OH− * | 2.43 ± 0.01 | 1.96 ± 0.00 | 1.94 ± 0.00 | 3.82 ± 0.02 | 1.87 ± 0.00 | 1.84 ± 0.00 | 3.11 ± 0.01 | 1.74 ± 0.00 |
| RN1 | 4.58 ± 0.05 | 1.92 ± 0.00 | 1.91 ± 0.00 | 3.82 ± 0.01 | 1.83 ± 0.00 | 1.84 ± 0.00 | 3.68 ± 0.03 | |
| RN2 & OH− | 2.45± 0.08 | 1.97 ± 0.01 | 1.90 ± 0.00 | 3.22±0.04 | 1.91 ± 0.01 | 1.85 ± 0.00 | 3.15 ± 0.03 | 1.72 ± 0.00 |
| RN2 | 2.26 ± 0.02 | 1.91 ± 0.00 | 1.87 ± 0.00 | 1.93 ± 0.00 | 1.88 ± 0.00 | 1.82 ± 0.00 | 3.43 ± 0.03 | |
| RN2-1 & OH− * | 2.41 ± 0.03 | 2.02 ± 0.01 | 1.92 ± 0.01 | 3.13 ± 0.03 | 1.90 ± 0.01 | 1.93 ± 0.01 | 3.38 ± 0.1 | 1.72 ± 0.01 |
| RN2-1* | 2.28 ± 0.02 | 1.95 ± 0.02 | 1.97 ± 0.03 | 1.98 ± 0.02 | 3.57 ± 0.01 | 1.87 ± 0.02 | 3.77 ± 0.04 | |
| RN3 & OH− | 2.44 ± 0.02 | 1.95 ± 0.01 | 1.90 ± 0.00 | 1.92 ± 0.00 | 3.22 ± 0.03 | 1.85 ± 0.00 | 3.15 ± 0.01 | 1.73 ± 0.00 |
| RN3 | 2.30 ± 0.02 | 1.91 ± 0.00 | 1.87 ± 0.00 | 1.94 ± 0.01 | 1.88 ± 0.00 | 1.82 ± 0.00 | 3.40 ± 0.02 | |
| RN3-1 & OH− * | 2.48 ± 0.01 | 2.00 ± 0.01 | 1.90 ± 0.01 | 1.90 ± 0.02 | 3.12 ± 0.04 | 1.91 ± 0.01 | 3.27 ± 0.05 | 1.72 ± 0.00 |
| RN3-1* | 2.31 ± 0.01 | 1.98 ± 0.00 | 1.89 ± 0.01 | 2.00 ± 0.01 | 3.49 ± 0.02 | 1.93 ± 0.01 | 3.80 ± 0.02 | |
Table 2.
Distance between Mg2+ ions and metal bridging hydroxide oxygen atom.
| Metal A - metal B (Å) | Metal A – O atom (Å) | Metal B – O atom (Å) | |
|---|---|---|---|
| Experimental | |||
| Nor-NOHA (3KV2) | 3.3 | 2.0a | 2.3a |
| Simulation | |||
| nor-NOHA* | 3.27 ± 0.02 | 1.90 ± 0.01a | 1.83 ± 0.01a |
| Arg+ OH | 3.05 ± 0 | 1.74 ± 0 | 1.73 ± 0 |
| Arg+ | 4.25 ± 0.01 | NA | NA |
| RN1 OH* | 3.15 ± 0 | 1.74 ± 0 | 1.74 ± 0 |
| RN1 | 5.14 ± 0.03 | NA | NA |
| RN2 OH | 3.06 ± 0.01 | 1.74 ± 0.01 | 1.72 ± 0 |
| RN2 | 4.48 ± 0.02 | NA | NA |
| RN2-1 OH* | 3.09 ± 0.03 | 1.79 ± 0.01 | 1.72 ± 0.01 |
| RN2-1* | 4.33 ± 0.02 | NA | NA |
| RN3 OH | 3.06 ± 0.02 | 1.74 ± 0.01 | 1.73 ± 0 |
| RN3 | 4.46 ± 0.01 | NA | NA |
| RN3-1 OH* | 3.08 ± 0.02 | 1.80 ± 0.01 | 1.72 ± 0 |
| RN3-1* | 4.25 ± 0.04 | NA | NA |
20ns simulation; NA: Not applicable as the metal bridging oxygen of the hydroxide is not present
The N(ω) hydroxy oxygen is a metal bridging atom in nor-NOHA case.
Additional criteria to validate the simulation protocol are hydrogen bonds by nor-NOHA with the surrounding protein environment. Experimentally nor-NOHA forms hydrogen bonds with Asp128, Asn130, Ser137, Asp183 and Thr246. The simulation captures all the hydrogen bonds observed experimentally, although the % of occurrence is low in certain cases. Table 3 shows the hydrogen bond data, where the % of occurrence of the observed hydrogen bonds is presented using a cutoff distance of 2.4 Å. The hydrogen bond with Asn130 (Hδ2) does not appear for more than 0.5% of the simulation due to a water molecule interacting frequently with the terminal carboxylate oxygens (OT1 and OT2) of nor-NOHA (Figure S3). Moreover, terminal carboxylate and amino groups mostly interact with protein atoms by means of water-mediated hydrogen bonds. Overall, the results of the nor-NOHA-arginase I simulation indicate that the use of Mg2+ parameters to model the Mn2+ ions in the active site is appropriate as well as indicate the ability of the applied simulation protocol to satisfactorily model the highly charged active site of arginase I.
Table 3.
Critical hydrogen bonds and their percent occurence between the protein and nor-NOHA. % occurrence was calculated based on a X…H cutoff distance of 2.4 Å.
| Res. id. | Protein atom |
nor-NOHA atom |
% of occurrence |
Experimental H-bond distance |
|---|---|---|---|---|
| Asp-128 | OD | HH12 | 69% | 1.8 |
| Asp-183 | OD | HT | 6% | 1.8 |
| Asn-130 | HD2 | OT2 | 0.5% | 2.3 |
| Ser-137 | HG1 | OT | 12% | 2.6 |
| Thr-246 | OG1 | HH21/HD | 1% | 2.1 |
Stability of substrate in the active site
Subsequent analysis focused on understanding the impact of the protonation and tautomerization state on the interaction of the Arg substrate with the arginase active site. To investigate the stability of the different forms of Arg in the active site the distances between the Mg2+ ions and the guanidium nitrogens (NH1 & NH2) were analyzed (Figure 5). For positive Arg, the guanidinium stays within 4 Å of the bi-metal center in both the presence and absence of the hydroxide ion. The neutral forms of Arg showed the presence of the hydroxide to have a large impact. In the absence of the hydroxide the guanidinium to bi-metal center distances stayed within 4 Å throughout the simulations. However, inclusion of the hydroxide destabilized the neutral substrates binding orientations in the active site, with the RN1 and RN3 tautomers starting to diffuse out of the pocket as indicated by the large increase in the substrate to Mg2+ distances (Figure 5) while RN2 moves up to 5 Å away from the Mg2+ ions. Thus, despite the highly charged bi-metal center the charged Arg can maintain a position in the active site such that it can undergo catalysis. This is maintained in either the presence or absence of the hydroxide ions. Contributing to this are additional Arg-protein interactions, as detailed below. In contrast, the neutral Arg tautomers are not stable in the active site, especially in the presence of the negatively charge hydroxide ion. This suggests a subtle balance of electrostatic interactions within the active site due to the presence of the positively charged metal ions, negatively charged amino acid side chains, charge/neutral substrate and presence or absence of the hydroxide ion (see Supplementary Figure S4). This balance appears to be perturbed if the substrate assumes a neutral state, regardless of the tautomerization state, and while they are more stable if a hydroxide is not present, the present results suggest that the neutral state of Arg is not involved in catalysis.
Figure 5.
Distance between substrate/inhibitor and magnesium ions as a function of simulation time.
Interestingly, the inhibitor nor-NOHA stays in the pocket throughout the simulation with an average guanidinium to bi-metal center distance of ~2.3 Å. This is interesting as the overall charge of nor-NOHA is neutral. However, it contains an additional oxygen atom on the guanidinium, yielding a zwitterion (Figure S3), which mimics the catalytic hydroxide. The stability of this inhibitor in the binding pocket, with an experimental Kd value of 51 nM, indicates that inhibitors mimicking the hydroxide ion can improve affinity. Its stability is also consistent with the ability of charged Arg in the presence of the hydroxide to be stabilized in the active site. Further analysis will, accordingly, focus on the structure of the active site in the presence of Arg+.
Structure of the Arginase Active Site
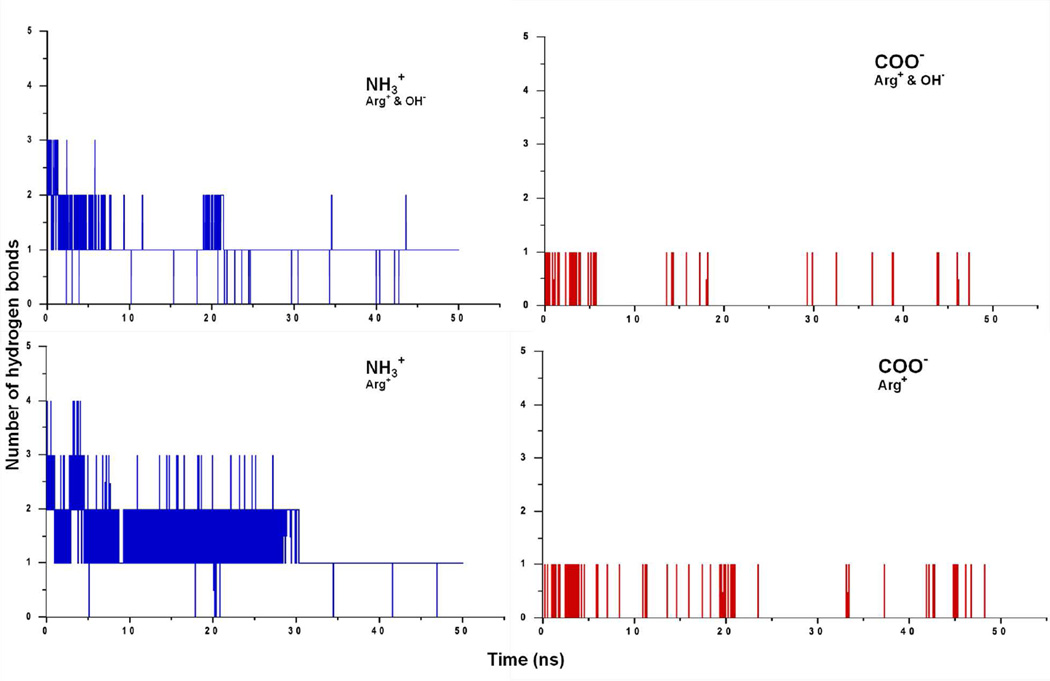
The arginase I substrate binding pocket is dominated by hydrophilic residues with the protein-substrate interactions mainly governed by electrostatic interactions. Additionally, there are several hydrogen bonds involving the guanidinium, α-amino and α-carboxylate groups, which further enhance the interaction betwen protein and substrate. Table 4 summarizes the arginine-protein interactions both in the presence and absence of hydroxide. The guanidinium nitrogens act as hydrogen bond donors to Asp128, Glu277, His141 and Thr246. The α-carboxylate group accepts hydrogen bonds from Asn130 and Ser137. The hydrogen bonds observed in the nor-NOHA and NOHA crystal structures suggest that Asn130-inhibitor interactions contribute to the binding of nor-NOHA,20 supporting the importance of this residue. The substrate α-amino group frequently interacts with Asp181, Asp183 and Glu186, though at a lower frequency than the other interactions due to the α-amino group interacting with water more often than α-carboxylate moiety. Figure 6 shows the number of hydrogen bonds established with water molecules during the simulations. There are several hydrogen bonds established by the amino group and at least one is maintained throughout the 50 ns simulations, with a tendency for more hydrogen bonds to be present in the absence of the hydroxide ion. All of the reported arginase I high affinity inhibitors have an α-amino and α-carboxylate moiety, indicating the essential role of these groups. Supporting this are MD results as part of this study showing that replacement of the carboxylate of Arg+ with a methylester leads to binding mode change (data not shown). Cama et al. demonstrated that deletion of these groups diminishes the binding affinity of nor-NOHA and 2(S)-amino-6-boronohexanoic acid (ABH).35 These results support the role of the charged carboxylate and amino moieties in strong inhibitor binding.
Table 4.
Hydrogen bonding between arginase I residues and charged arginine.
| Res. Id. | Protein atom |
substrate atom |
% occurrence | |
|---|---|---|---|---|
| Arg+ & OH− |
Arg+ | |||
| Donor | Acceptor | |||
| Asn130 | ND2 | OT1 | 28.02 | 22.18 |
| Thr135 | OG1 | OT1 | 0.01 | NA |
| Thr136 | OG1 | OT1 | 0.01 | NA |
| Ser137 | OG | OT1 | 16.39 | 7.57 |
| Ser137 | OG | OT2 | 61.42 | 37.2 |
| Asn139 | ND2 | OT2 | 0.27 | 0.05 |
| Acceptor | Donor | Arg+ & OH− |
Arg+ | |
| Asp128 | OD | NE | 101.57 | 31.53 |
| Asp128 | OD | NH1 | 20.62 | 11.47 |
| His141 | O | NH1 | 94.12 | 36.48 |
| His141 | O | NE | 0.01 | 0.17 |
| Asp181 | OD | N | 19.21 | NA |
| Asp183 | OD | N | 5.26 | 40.8 |
| Glu186 | OE | N | 14.91 | 0.69 |
| Asp232 | OD | NH1 | NA | 0.14 |
| Thr246 | OG1 | NH2 | 97.25 | 27.89 |
| Glu277 | OE | NH1 | 99.98 | 40.19 |
| Glu277 | OE | NH2 | 71.13 | 47.91 |
NA: No hydrogen bond observed.
Figure 6.
Number of hydrogen bonds between water and the α-amino and α-caboxylate groups of the charged arginine substrate. Upper and lower panel corresponding to with and without hydroxide ions, respectively.
The key role of the metal ions is to activate the hydroxide ion17 by creating a strong electrostatic field inside the binding site. The Arg+-hydroxide simulation reveals that the metal ions Mg2+A and Mg2+B maintain the experimentally determined positions of the manganese ions,36 Mn2+A – Mn2+B in the presence of hydroxide ion, which acts to bridge the metals (Figure 7). The experimental distance between the inhibitor oxygen (Figure 1) and the metal (Mn2+) is 3.3 Å in the nor-NOHA-Arginase I crystal structure. Similarly, in the Arg+-hydroxide simulation a distance of 3.05 Å is sampled (Table 2) indicating the structure of the cluster is being maintained. There is a considerable increase in the Mg2+A – Mg2+B distance to 4.25 Å in the Arg+ alone simulation, which disrupts the structure of the cluster. These results indicate that the hydroxide ion acts to bridge the metal ions indicating its importance in the maintaining a functional metal cluster of Arginase I.
Figure 7.
Bi-metal cluster of arginase I. Distances correspond to block averages from the Arg+ - hydroxide simulation. Mg+2 ions A and B are shown as green spheres and the oxygen of the hydroxide is shown as a red sphere.
Metal ion to side chain distances obtained from the Arg+-hydroxide simulation are shown in Figure 7. The Mg2+ to protein atom distances are in good agreement with those in the crystal structure of nor-NOHA-arginase I (Table 1) and from other arginase I crystal structures.20 From the geometry of the active site it is clear that a balance of ionic interaction, including that of the hydroxide ion, which serves as a nucleophile in the catalyzed hydrolysis of arginine by arginase,17 stabilize the structure of the active site as required for catalysis to occur.
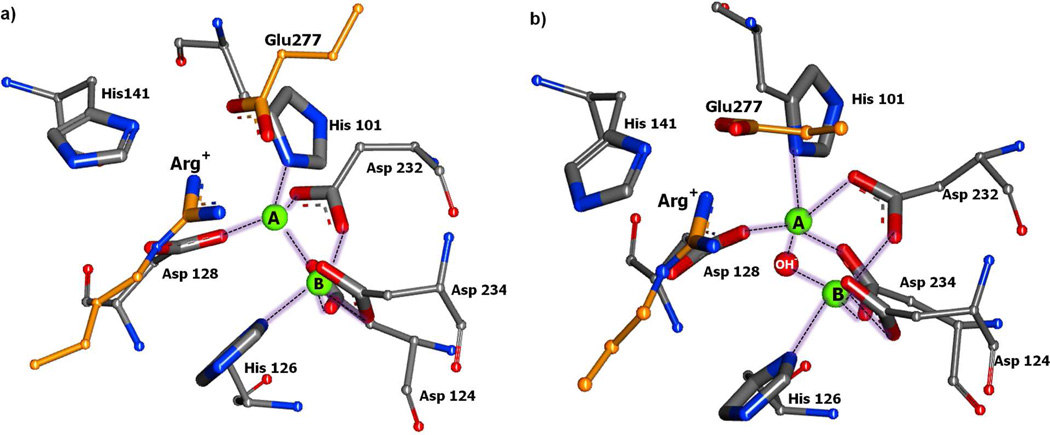
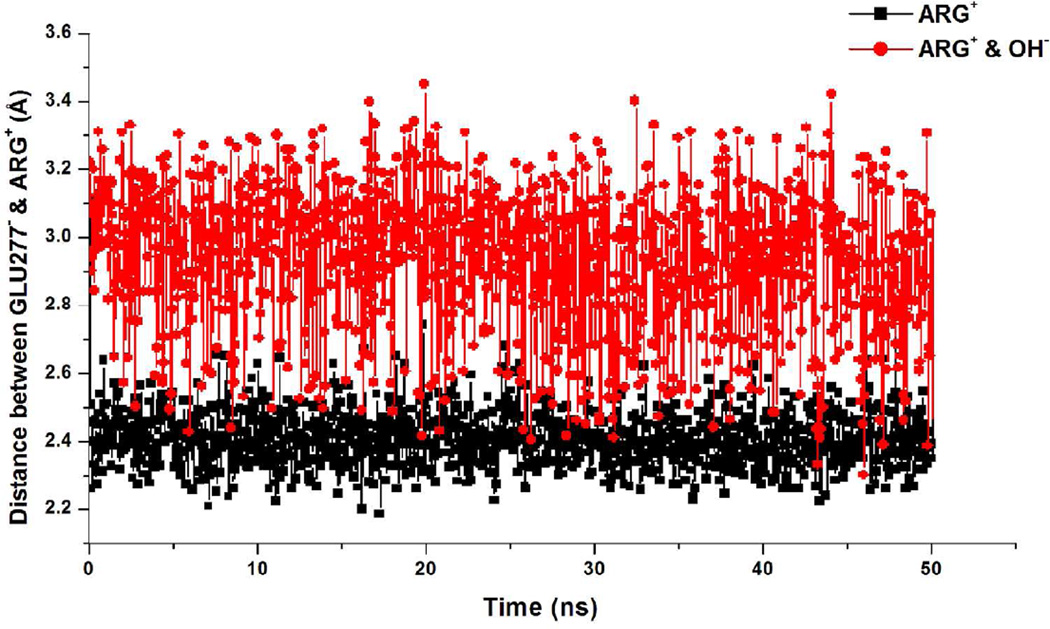
Glu277 has been shown to be essential for the catalytic activity of arginase I.37 In the absence of substrate Glu277 hydrogen bonds with His14130 while in the presence of substrate there is an ionic interaction between the carboxylate of Glu277 and the guanidinium of Arg (Figure 8). This interaction is much more favorable in the absence of the hydroxide ion (Figures 8A and 9), while in the presence of the hydroxide the Glu277-Arg interaction is perturbed and there is a direct interaction between the guanidinium and the hydroxide (Figure 8B). In addition, a crystal structure of human apo arginase I (PDB:2ZAV)36 reveals three water molecules forming a H-bond network in the active site. The presence of these water, one of which may be a hydroxide, has been suggested to indicate that deprotonation of a water is facilitated by proton transfer with His141 acting as a proton shuttle.38 This data is consistent with a proposed role of a metal-bridging water molecule in balancing ionic interactions when Arg+ initially binds in the active site. In our simulations no water was present adjacent to the bi-metal cluster in the Arg+ simulation without the hydroxide, which could be due to the solvent overlay procedure and the inability for water to diffuse into the bottom of the active site on the time scale of the simulations. However, assuming that a water molecule may be located in this region in the absence of the hydroxide as indicated by the crystal study, these observations suggest that when the substrate initially binds Glu277 is critical for organizing the constituents of the active site to facilitate ionization of the water leading to the formation of the hydroxide. Once formed, the presence of the hydroxide leads to the Glu277 interaction becoming less favorable (Figure 9), consistent with the additional negative charge, but still acting to position the scissile carbon of the guanidinium moiety over hydroxide ion thereby facilitating nucleophilic attack.
Figure 8.
Active site of Arginase I showing how the absence (a) or presence (b) of the hydroxide ion affects the salt bridge interaction between Arg+ and Glu 277. Metal coordinating residues shown in ball and stick with metal (green sphere) and hydroxide (red sphere), susbtrate and Glu277 in orange stick without backbone atoms. Hydrogen atoms are not shown for the sake of clarity. The image is from the 2 ns snapshot from the respective MD simulations.
Figure 9.
Salt-bridge interaction plotted as distance between geometric center of nitrogens and oxygens of Arg+ and Glu277, respectively.
Conclusion
MD simulation studies were used to elucidate the charge dependent behavior of Arg+/0 in the arginase I active site. Results indicate that Arg is charged in the arginase I active site as the protonated group is critical for stabilizing the substrate in the binding site in both the presence and absence of a hydroxide ion. This stabilization is, in part due to a salt bridge with Glu277, which is hypothesized to be critical for orienting the guanidinium of the substrate in an orientation that facilitates ionization of a water molecule leading to formation of a hydroxide ion. Glu277 also helps in positioning the guanidinium of the substrate in an orientation that allow for nucleophilic attack by the hydroxide rather than directly coordinating with the active site metals.
Concerning inhibitor design, the present results suggest features that could enhance the affinity of an arginase I inhibitor. For example, an inhibitor may mimic the coordination of the bi-metal cluster that occurs with Arg0. Known inhibitors that act in this way and include an atom with high electronegativity (eg. oxygen) on the moiety that is involved in inner sphere coordination have nanomolar affinities.30 Inhibitors could be designed to mimic the features of the substrate. Such inhibitors would contain a moiety capable of expelling the proposed metal coordinating water, and therefore not be able to undergo nucleophilic attack as well as have strong binding affinities. Such Arginase I inhibitors may closely mimic the transition state in the active site without being hydrolyzed do to the omission of the water. Moreover, retaining the α-amino and α-carboxylate moieties is a key requirement in inhibitors of this target.
Supplementary Material
ACKNOWLEDGMENT
The authors acknowledge support from the University of Maryland Computer-Aided Drug Design Center, Accelrys Inc. for providing access to Discovery Studio and the NIH (GM072558).
Glossary
ABBREVIATIONS
- NOS
nitric oxide synthase
- NO
nitric oxide
- nor-NOHA
Nω-hydroxy-L-nor-arginine
- NOHA
Nω-hydroxy-L-arginine
- MD
MOlecular Dynamics
- RMSD
Root mean square deviation
Footnotes
Supporting Information. Details of simulation system composition and time scale, nor-NOHA RMSF data for the 20 ns simulation and nor-NOHA structure with atom label. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Iyer R, Jenkinson CP, Vockley JG, Kern RM, Grody WW, Cederbaum S. The human arginases and arginase deficiency. J. Inherit. Metab. Dis. 1998;21(Suppl 1):86–100. doi: 10.1023/a:1005313809037. [DOI] [PubMed] [Google Scholar]
- 2.Christianson DW. Arginase: structure, mechanism, and physiological role in male and female sexual arousal. Acc. Chem. Res. 2005;38:191–201. doi: 10.1021/ar040183k. [DOI] [PubMed] [Google Scholar]
- 3.Morris SM., Jr Regulation of enzymes of the urea cycle and arginine metabolism. Annu. Rev. Nutr. 2002;22:87–105. doi: 10.1146/annurev.nutr.22.110801.140547. [DOI] [PubMed] [Google Scholar]
- 4.Gotoh T, Araki M, Mori M. Chromosomal localization of the human arginase II gene and tissue distribution of its mRNA. Biochem. Biophys. Res. Commun. 1997;233:487–491. doi: 10.1006/bbrc.1997.6473. [DOI] [PubMed] [Google Scholar]
- 5.Bowcutt R, Bell LV, Little M, Wilson J, Booth C, Murray PJ, Else KJ, Cruickshank SM. Arginase-1-expressing macrophages are dispensable for resistance to infection with the gastrointestinal helminth Trichuris muris. Parasite Immunol. 2011;33:411–420. doi: 10.1111/j.1365-3024.2011.01300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nieves C, Jr, Langkamp-Henken B. Arginine and immunity: a unique perspective. Biomed. Pharmacother. 2002;56:471–482. doi: 10.1016/s0753-3322(02)00291-3. [DOI] [PubMed] [Google Scholar]
- 7.Rodriguez PC, Quiceno DG, Zabaleta J, Ortiz B, Zea AH, Piazuelo MB, Delgado A, Correa P, Brayer J, Sotomayor EM, Antonia S, Ochoa JB, Ochoa AC. Arginase I production in the tumor microenvironment by mature myeloid cells inhibits T-cell receptor expression and antigen-specific T-cell responses. Cancer Res. 2004;64:5839–5849. doi: 10.1158/0008-5472.CAN-04-0465. [DOI] [PubMed] [Google Scholar]
- 8.Cama E, Colleluori DM, Emig FA, Shin H, Kim SW, Kim NN, Traish AM, Ash DE, Christianson DW. Human arginase II: crystal structure and physiological role in male and female sexual arousal. Biochemistry. 2003;42:8445–8451. doi: 10.1021/bi034340j. [DOI] [PubMed] [Google Scholar]
- 9.Maarsingh H, Zaagsma J, Meurs H. Arginase: a key enzyme in the pathophysiology of allergic asthma opening novel therapeutic perspectives. Br. J. Pharmacol. 2009;158:652–664. doi: 10.1111/j.1476-5381.2009.00374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benson RC, Hardy KA, Morris CR. Arginase and arginine dysregulation in asthma. J. Allergy (Cairo) 2011;2011 doi: 10.1155/2011/736319. 736319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang J, George SC. Modeling gas phase nitric oxide release in lung epithelial cells. Nitric Oxide. 2011;25:275–281. doi: 10.1016/j.niox.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bagnost T, Ma L, da Silva RF, Rezakhaniha R, Houdayer C, Stergiopulos N, Andre C, Guillaume Y, Berthelot A, Demougeot C. Cardiovascular effects of arginase inhibition in spontaneously hypertensive rats with fully developed hypertension. Cardiovasc. Res. 2010;87:569–577. doi: 10.1093/cvr/cvq081. [DOI] [PubMed] [Google Scholar]
- 13.Zhou L, Zhu DY. Neuronal nitric oxide synthase: structure, subcellular localization, regulation, and clinical implications. Nitric Oxide. 2009;20:223–230. doi: 10.1016/j.niox.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Dowling DP, Ilies M, Olszewski KL, Portugal S, Mota MM, Llinas M, Christianson DW. Crystal structure of arginase from Plasmodium falciparum and implications for L-arginine depletion in malarial infection. Biochemistry. 2010;49:5600–5608. doi: 10.1021/bi100390z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iniesta V, Gomez-Nieto LC, Corraliza I. The inhibition of arginase by N(omega)-hydroxy-l-arginine controls the growth of Leishmania inside macrophages. J. Exp. Med. 2001;193:777–7784. doi: 10.1084/jem.193.6.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pokrovskiy MV, Korokin MV, Tsepeleva SA, Pokrovskaya TG, Gureev VV, Konovalova EA, Gudyrev OS, Kochkarov VI, Korokina LV, Dudina EN, Babko AV, Terehova EG. Arginase inhibitor in the pharmacological correction of endothelial dysfunction. Int. J. Hypertens. 2011;2011 doi: 10.4061/2011/515047. 515047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christianson DW, Cox JD. Catalysis by metal-activated hydroxide in zinc and manganese metalloenzymes. Annu. Rev. Biochem. 1999;68:33–57. doi: 10.1146/annurev.biochem.68.1.33. [DOI] [PubMed] [Google Scholar]
- 18.Cama E, Emig FA, Ash DE, Christianson DW. Structural and functional importance of first-shell metal ligands in the binuclear manganese cluster of arginase I. Biochemistry. 2003;42:7748–7758. doi: 10.1021/bi030074y. [DOI] [PubMed] [Google Scholar]
- 19.Ilies M, Di Costanzo L, North ML, Scott JA, Christianson DW. 2-aminoimidazole amino acids as inhibitors of the binuclear manganese metalloenzyme human arginase I. J. Med. Chem. 2010;53:4266–4276. doi: 10.1021/jm100306a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Di Costanzo L, Ilies M, Thorn KJ, Christianson DW. Inhibition of human arginase I by substrate and product analogues. Arch. Biochem. Biophys. 2010;496:101–108. doi: 10.1016/j.abb.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall NF, Sprinkle MR. Relations between the structure and strength of certain organic bases in aqueous solution. J. Am. Chem. Soc. 1932;54:3469–3485. [Google Scholar]
- 22.Angyal SJ, Warburton WK. The basic strengths of methylated guanidines. J. Chem. Soc. 1951;1951:2492–2494. [Google Scholar]
- 23.Nozaki Y, Tanford C. Examination of titration behavior. Methods Enzymol. 1967;11:715–734. [Google Scholar]
- 24.Brooks BR, Brooks CL, 3rd, Mackerell AD, Jr, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. CHARMM: the biomolecular simulation program. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buck M, Bouguet-Bonnet S, Pastor RW, MacKerell AD., Jr Importance of the CMAP correction to the CHARMM22 protein force field: dynamics of hen lysozyme. Biophys. J. 2006;90:L36–L38. doi: 10.1529/biophysj.105.078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, Mackerell AD., Jr CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li L, Vorobyov I, Allen TW. Potential of Mean Force and pKa Profile Calculation for a Lipid Membrane-Exposed Arginine Side Chain. J. Phys. Chem. B. 2008;112:9574–9587. doi: 10.1021/jp7114912. [DOI] [PubMed] [Google Scholar]
- 29.Li L, Vorobyov I, MacKerell AD, Allen TW. Is Arginine Charged in a Membrane? Biophys. J. 2008;94:L11–L13. doi: 10.1529/biophysj.107.121566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Di Costanzo L, Sabio G, Mora A, Rodriguez PC, Ochoa AC, Centeno F, Christianson DW. Crystal structure of human arginase I at 1.29-A resolution and exploration of inhibition in the immune response. Proc. Natl. Acad. Sci. U. S. A. 2005;102:13058–13063. doi: 10.1073/pnas.0504027102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Word JM, Lovell SC, Richardson JS, Richardson DC. Asparagine and glutamine: using hydrogen atom contacts in the choice of side-chain amide orientation. J. Mol. Biol. 1999;285:1735–1747. doi: 10.1006/jmbi.1998.2401. [DOI] [PubMed] [Google Scholar]
- 32.Jo S, Kim T, Iyer VG, Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 33.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 34.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 35.Cama E, Pethe S, Boucher JL, Han S, Emig FA, Ash DE, Viola RE, Mansuy D, Christianson DW. Inhibitor coordination interactions in the binuclear manganese cluster of arginase. Biochemistry. 2004;43:8987–8299. doi: 10.1021/bi0491705. [DOI] [PubMed] [Google Scholar]
- 36.Di Costanzo L, Pique ME, Christianson DW. Crystal structure of human arginase I complexed with thiosemicarbazide reveals an unusual thiocarbonyl mu-sulfide ligand in the binuclear manganese cluster. J. Am. Chem. Soc. 2007;129:6388–6389. doi: 10.1021/ja071567j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lipscomb WN, Strater N. Recent Advances in Zinc Enzymology. Chem. Rev. 1996;96:2375–2434. doi: 10.1021/cr950042j. [DOI] [PubMed] [Google Scholar]
- 38.Kanyo ZF, Scolnick LR, Ash DE, Christianson DW. Structure of a unique binuclear manganese cluster in arginase. Nature. 1996;383:554–557. doi: 10.1038/383554a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.