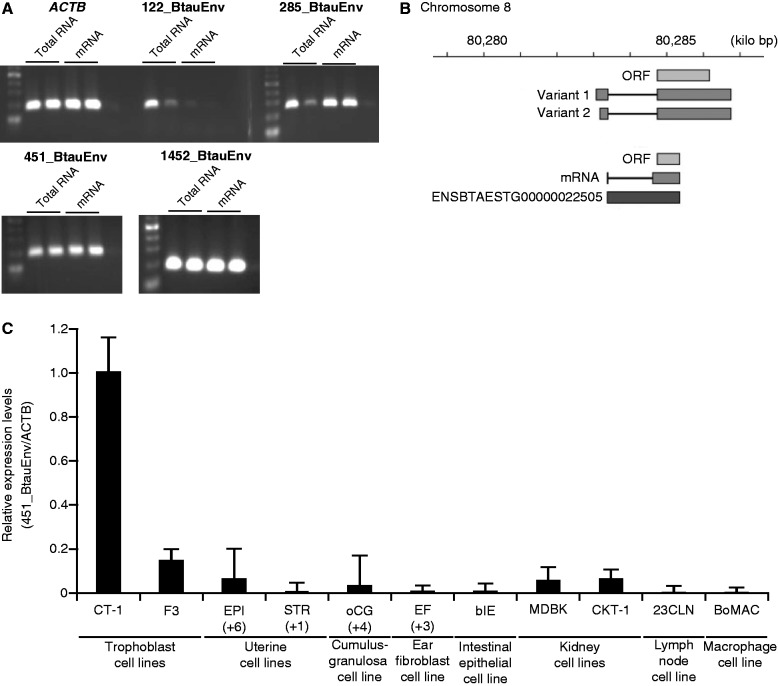
Fig. 3.—
Validation of env-derived sequences detected by SOLiD sequencing. Transcripts of env-derived genes were validated by PCR (A). Total RNA (1,000 ng, n = 2 each) or poly(A)+ RNA (30 ng, n = 2 each), isolated from CT-1 cells, was reverse transcribed and subjected to PCR analysis with primers presented in supplementary table S8, Supplementary Material online. ACTB mRNA was used as an internal control. (B) Chromosomal locations of BERV-P mRNA variants. 5′- and 3′-RACE was used to clone the entire BERV-P cDNA. There are two variants of BERV-P, resulting from difference in lengths of 5′-noncoding regions. BERV-P transcripts were found in various bovine cell lines and primary cells (C). Total RNAs (1,000 ng, n = 2 each), isolated from trophoblast cell lines (CT-1 or F3), endometrial epithelial (EPI) or stromal (STR) primary cells, ovarian cumulus granulosa (oCG) primary cells, ear-derived fibroblast (EF) primary cells, intestinal epithelial cell line (bIE), kidney cell lines (MDBK or CKT-1), lymph node cell line (23CLN), or macrophage cell line (BoMAC), were subjected to real-time PCR for BERV-P transcripts with primers presented in supplementary table S8, Supplementary Material online. ACTB transcript was used as endogenous reference gene product. The expression of the transcript was normalized to the expression of ACTB measured in the same cDNA preparation. The values are shown as the mean ± standard error of the mean.