Fig. 6.—
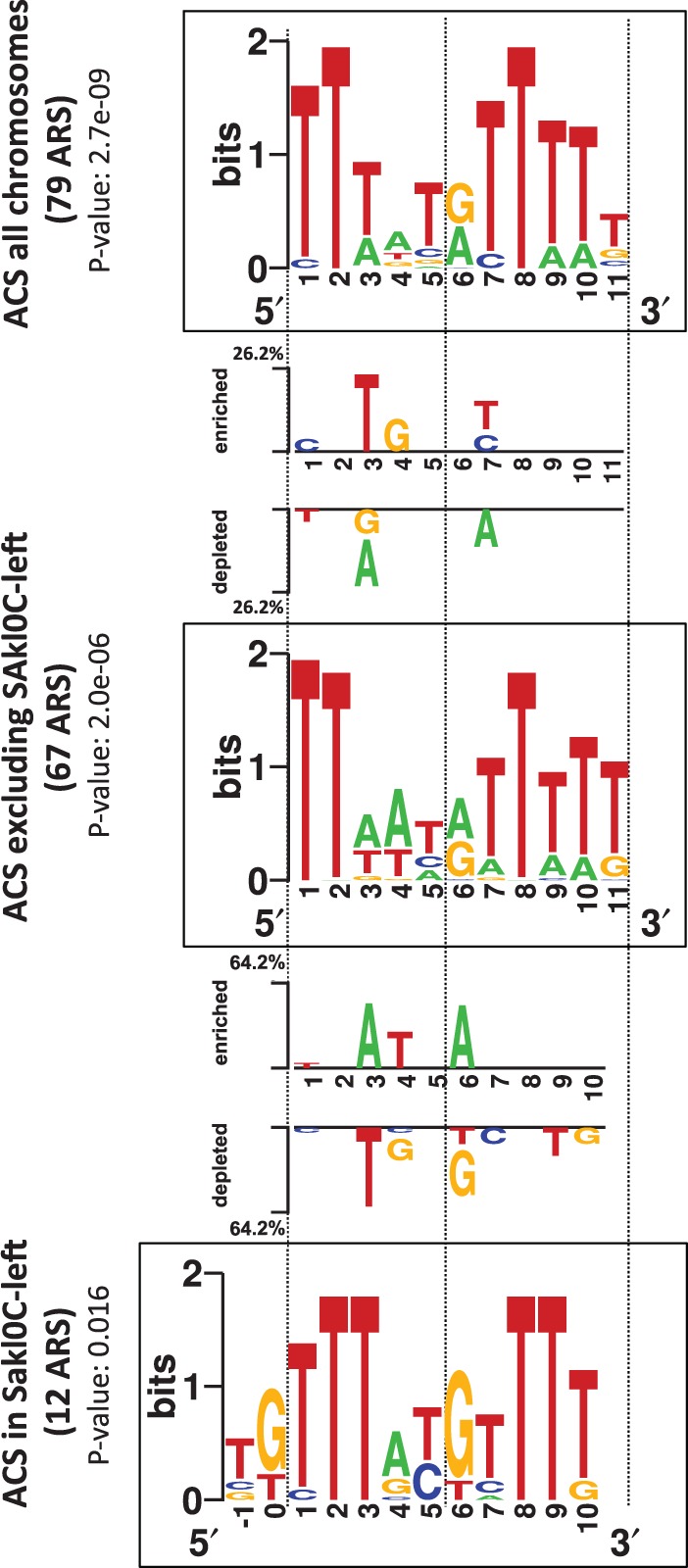
Identification of different ACS motifs in the genome of Lachancea kluyveri. The three squared logo sequences correspond to putative ACS motifs for all chromosomes (top), for all chromosomes excluding Sakl0C-left (middle), and for Sakl0C-left (bottom). The most significant motif for Sakl0C-left was obtained for a width of 12 bp instead of 11 bp for all other chromosomes and shifted by 2 bp to the 5'-end of the 11 bp motif. Significant base differences between “ACS of all chromosomes” (top) and “ACS excluding Sakl0C-left” (middle), and between “ACS excluding Sakl0C-left” (middle) and “ACS of Sakl0C-left” (bottom) are represented between the corresponding squared logos with the indication of the enriched or depleted bases at each position.
