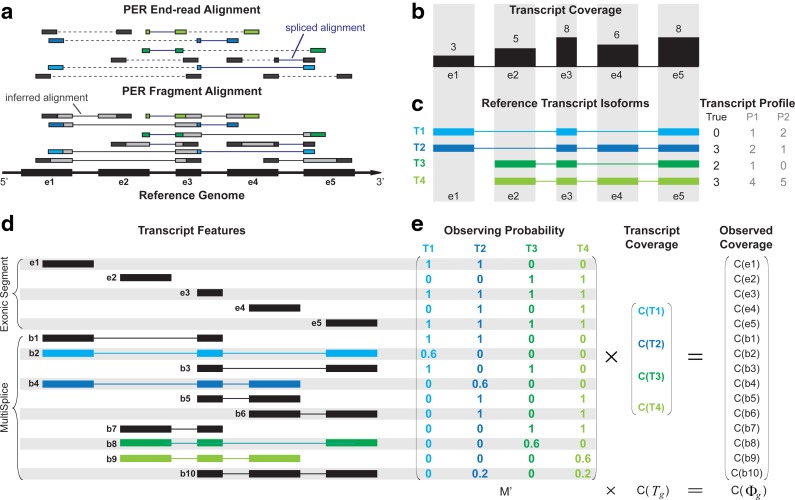
FIG. 1.
Overview of the MultiSplice model. (a) Sequenced RNA-seq short-reads are first mapped to the reference genome using an RNA-seq read aligner such as MapSplice (Wang et al., 2010). In the presence of paired-end reads, MapPER (Hu et al., 2010) can be applied to find PER fragment alignments for the entire transcript fragment based on the distribution of insert size. (b) Observed coverage on each exonic segment. (c) Four transcripts originate from the alternative start and exon-skipping events. Provided with these transcripts, abundance estimates would be unidentifiable for methods that only use coverage on exonic segments. Both transcript profiles P1 and P2, for instance, can explain the observed read coverage on each exon but deviate from the true transcript expression profile. (d) MultiSplices that connect multiple exonic segments in a transcript. (e) A linear model can be set up where the expected coverage on every exonic or MultiSplice feature approximates its observed coverage. The transcript expression is solved as the one that minimizes the sum of squared relative error.