Abstract
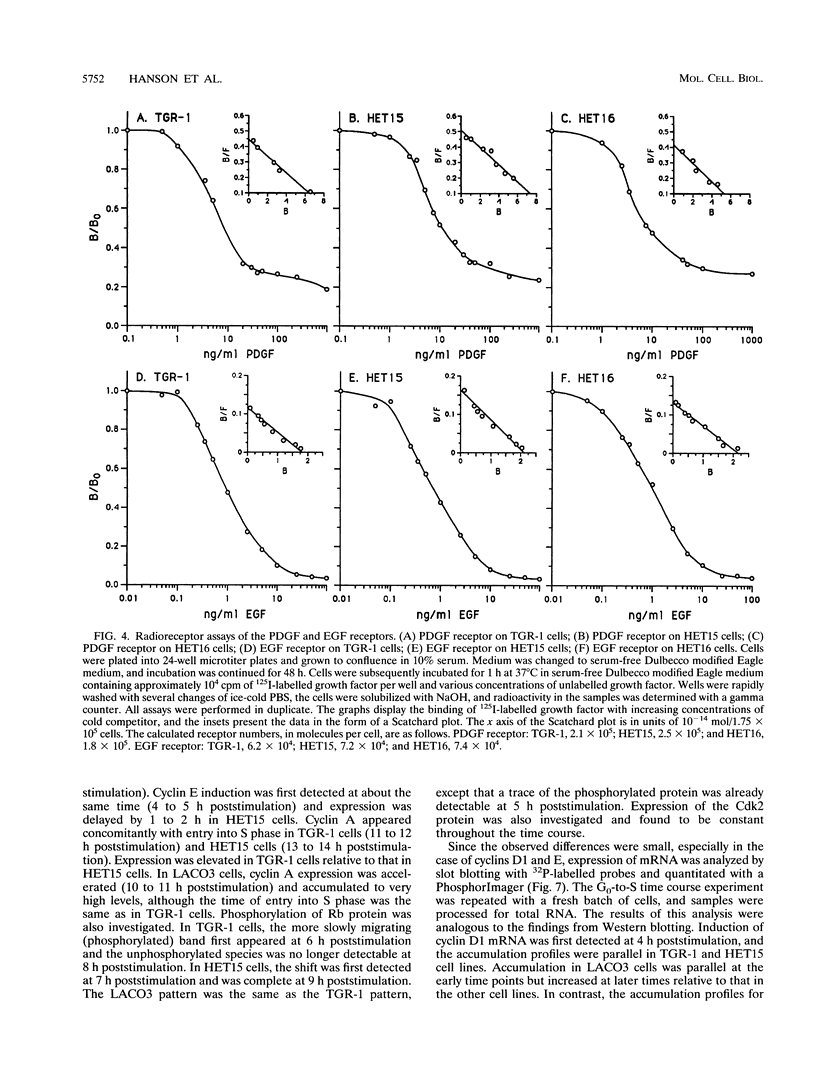
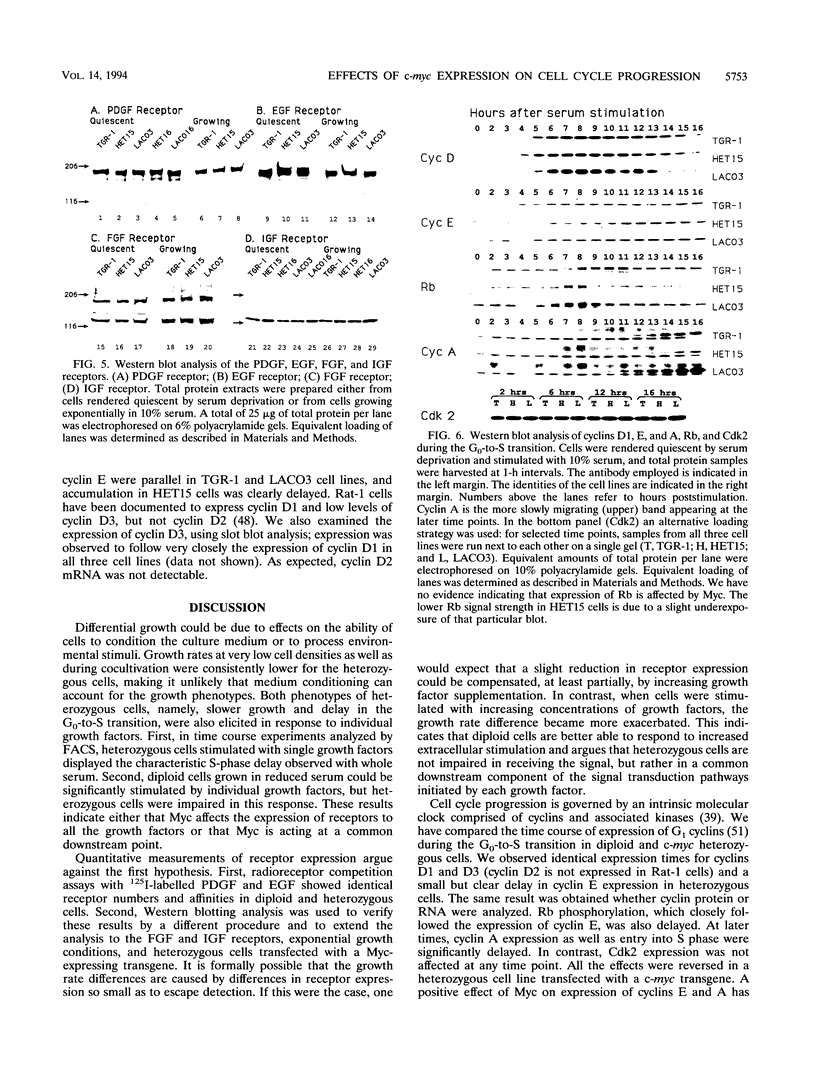
We used targeted homologous recombination to disrupt one c-myc gene copy in a diploid fibroblast cell line and found that a twofold reduction in Myc expression resulted in lower exponential growth rates and a lengthening of the G0-to-S-phase transition (M. Shichiri, K. D. Hanson and J. M. Sedivy, Cell Growth Differ. 4:93-104, 1993). Myc is a transcription factor, and the number of target genes whose regulation could result in differential growth rates may be very large. We have approached this problem by examining effects of reduced c-myc expression in three broad areas: (i) secretion of growth factors, (ii) expression of growth factor receptors, and (iii) intracellular signal transduction between Myc and components of the intrinsic cell cycle clock. We have found no evidence that differential medium conditioning can account for the growth phenotypes. Likewise, the expression of receptors for platelet-derived growth factor, epidermal growth factor, basic fibroblast growth factor, and insulin-like growth factor I was the same in diploid and heterozygous cells (platelet-derived growth factor, epidermal growth factor, fibroblast growth factor, and insulin-like growth factor are the sole growth factors required by these cells for growth in serum-free medium). In contrast, expression of cyclin E, cyclin A, and Rb phosphorylation were delayed when quiescent c-myc heterozygous cells were stimulated to enter the cell cycle. Expression of cyclin D1, cyclin D3, and Cdk2 was not affected. The timing of cyclin E induction was the earliest observable effect of reduced Myc expression. Our data indicate that Myc contributes to regulation of proliferation by a cell-autonomous mechanism that involves the modulation of cyclin E expression and, consequently, progression through the restriction point of the cell cycle.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baldin V., Lukas J., Marcote M. J., Pagano M., Draetta G. Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes Dev. 1993 May;7(5):812–821. doi: 10.1101/gad.7.5.812. [DOI] [PubMed] [Google Scholar]
- Beckmann H., Su L. K., Kadesch T. TFE3: a helix-loop-helix protein that activates transcription through the immunoglobulin enhancer muE3 motif. Genes Dev. 1990 Feb;4(2):167–179. doi: 10.1101/gad.4.2.167. [DOI] [PubMed] [Google Scholar]
- Benvenisty N., Leder A., Kuo A., Leder P. An embryonically expressed gene is a target for c-Myc regulation via the c-Myc-binding sequence. Genes Dev. 1992 Dec;6(12B):2513–2523. doi: 10.1101/gad.6.12b.2513. [DOI] [PubMed] [Google Scholar]
- Bishop J. M. Molecular themes in oncogenesis. Cell. 1991 Jan 25;64(2):235–248. doi: 10.1016/0092-8674(91)90636-d. [DOI] [PubMed] [Google Scholar]
- Blackwell T. K., Huang J., Ma A., Kretzner L., Alt F. W., Eisenman R. N., Weintraub H. Binding of myc proteins to canonical and noncanonical DNA sequences. Mol Cell Biol. 1993 Sep;13(9):5216–5224. doi: 10.1128/mcb.13.9.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell T. K., Kretzner L., Blackwood E. M., Eisenman R. N., Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science. 1990 Nov 23;250(4984):1149–1151. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- Blackwood E. M., Eisenman R. N. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991 Mar 8;251(4998):1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- Campisi J., Gray H. E., Pardee A. B., Dean M., Sonenshein G. E. Cell-cycle control of c-myc but not c-ras expression is lost following chemical transformation. Cell. 1984 Feb;36(2):241–247. doi: 10.1016/0092-8674(84)90217-4. [DOI] [PubMed] [Google Scholar]
- Carr C. S., Sharp P. A. A helix-loop-helix protein related to the immunoglobulin E box-binding proteins. Mol Cell Biol. 1990 Aug;10(8):4384–4388. doi: 10.1128/mcb.10.8.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole M. D. The myc oncogene: its role in transformation and differentiation. Annu Rev Genet. 1986;20:361–384. doi: 10.1146/annurev.ge.20.120186.002045. [DOI] [PubMed] [Google Scholar]
- Davis A. C., Wims M., Spotts G. D., Hann S. R., Bradley A. A null c-myc mutation causes lethality before 10.5 days of gestation in homozygotes and reduced fertility in heterozygous female mice. Genes Dev. 1993 Apr;7(4):671–682. doi: 10.1101/gad.7.4.671. [DOI] [PubMed] [Google Scholar]
- Dean M., Levine R. A., Ran W., Kindy M. S., Sonenshein G. E., Campisi J. Regulation of c-myc transcription and mRNA abundance by serum growth factors and cell contact. J Biol Chem. 1986 Jul 15;261(20):9161–9166. [PubMed] [Google Scholar]
- Eilers M., Schirm S., Bishop J. M. The MYC protein activates transcription of the alpha-prothymosin gene. EMBO J. 1991 Jan;10(1):133–141. doi: 10.1002/j.1460-2075.1991.tb07929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan G. I., Wyllie A. H., Gilbert C. S., Littlewood T. D., Land H., Brooks M., Waters C. M., Penn L. Z., Hancock D. C. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992 Apr 3;69(1):119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- Ewen M. E., Sluss H. K., Whitehouse L. L., Livingston D. M. TGF beta inhibition of Cdk4 synthesis is linked to cell cycle arrest. Cell. 1993 Sep 24;74(6):1009–1020. doi: 10.1016/0092-8674(93)90723-4. [DOI] [PubMed] [Google Scholar]
- Field J. K., Spandidos D. A. The role of ras and myc oncogenes in human solid tumours and their relevance in diagnosis and prognosis (review). Anticancer Res. 1990 Jan-Feb;10(1):1–22. [PubMed] [Google Scholar]
- Fisher F., Goding C. R. Single amino acid substitutions alter helix-loop-helix protein specificity for bases flanking the core CANNTG motif. EMBO J. 1992 Nov;11(11):4103–4109. doi: 10.1002/j.1460-2075.1992.tb05503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor P. D., Sawadogo M., Roeder R. G. The adenovirus major late transcription factor USF is a member of the helix-loop-helix group of regulatory proteins and binds to DNA as a dimer. Genes Dev. 1990 Oct;4(10):1730–1740. doi: 10.1101/gad.4.10.1730. [DOI] [PubMed] [Google Scholar]
- Halazonetis T. D., Kandil A. N. Determination of the c-MYC DNA-binding site. Proc Natl Acad Sci U S A. 1991 Jul 15;88(14):6162–6166. doi: 10.1073/pnas.88.14.6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper J. W., Adami G. R., Wei N., Keyomarsi K., Elledge S. J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993 Nov 19;75(4):805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- Heikkila R., Schwab G., Wickstrom E., Loke S. L., Pluznik D. H., Watt R., Neckers L. M. A c-myc antisense oligodeoxynucleotide inhibits entry into S phase but not progress from G0 to G1. 1987 Jul 30-Aug 5Nature. 328(6129):445–449. doi: 10.1038/328445a0. [DOI] [PubMed] [Google Scholar]
- Holt J. T., Redner R. L., Nienhuis A. W. An oligomer complementary to c-myc mRNA inhibits proliferation of HL-60 promyelocytic cells and induces differentiation. Mol Cell Biol. 1988 Feb;8(2):963–973. doi: 10.1128/mcb.8.2.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T. Cooperation between oncogenes. Cell. 1991 Jan 25;64(2):249–270. doi: 10.1016/0092-8674(91)90637-e. [DOI] [PubMed] [Google Scholar]
- Inghirami G., Grignani F., Sternas L., Lombardi L., Knowles D. M., Dalla-Favera R. Down-regulation of LFA-1 adhesion receptors by C-myc oncogene in human B lymphoblastoid cells. Science. 1990 Nov 2;250(4981):682–686. doi: 10.1126/science.2237417. [DOI] [PubMed] [Google Scholar]
- Jansen-Dürr P., Meichle A., Steiner P., Pagano M., Finke K., Botz J., Wessbecher J., Draetta G., Eilers M. Differential modulation of cyclin gene expression by MYC. Proc Natl Acad Sci U S A. 1993 Apr 15;90(8):3685–3689. doi: 10.1073/pnas.90.8.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczmarek L., Hyland J. K., Watt R., Rosenberg M., Baserga R. Microinjected c-myc as a competence factor. Science. 1985 Jun 14;228(4705):1313–1315. doi: 10.1126/science.4001943. [DOI] [PubMed] [Google Scholar]
- Kato G. J., Barrett J., Villa-Garcia M., Dang C. V. An amino-terminal c-myc domain required for neoplastic transformation activates transcription. Mol Cell Biol. 1990 Nov;10(11):5914–5920. doi: 10.1128/mcb.10.11.5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato G. J., Lee W. M., Chen L. L., Dang C. V. Max: functional domains and interaction with c-Myc. Genes Dev. 1992 Jan;6(1):81–92. doi: 10.1101/gad.6.1.81. [DOI] [PubMed] [Google Scholar]
- Kelly K., Cochran B. H., Stiles C. D., Leder P. Cell-specific regulation of the c-myc gene by lymphocyte mitogens and platelet-derived growth factor. Cell. 1983 Dec;35(3 Pt 2):603–610. doi: 10.1016/0092-8674(83)90092-2. [DOI] [PubMed] [Google Scholar]
- Kerkhoff E., Bister K., Klempnauer K. H. Sequence-specific DNA binding by Myc proteins. Proc Natl Acad Sci U S A. 1991 May 15;88(10):4323–4327. doi: 10.1073/pnas.88.10.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyomarsi K., Sandoval L., Band V., Pardee A. B. Synchronization of tumor and normal cells from G1 to multiple cell cycles by lovastatin. Cancer Res. 1991 Jul 1;51(13):3602–3609. [PubMed] [Google Scholar]
- Koff A., Cross F., Fisher A., Schumacher J., Leguellec K., Philippe M., Roberts J. M. Human cyclin E, a new cyclin that interacts with two members of the CDC2 gene family. Cell. 1991 Sep 20;66(6):1217–1228. doi: 10.1016/0092-8674(91)90044-y. [DOI] [PubMed] [Google Scholar]
- Lassar A. B., Buskin J. N., Lockshon D., Davis R. L., Apone S., Hauschka S. D., Weintraub H. MyoD is a sequence-specific DNA binding protein requiring a region of myc homology to bind to the muscle creatine kinase enhancer. Cell. 1989 Sep 8;58(5):823–831. doi: 10.1016/0092-8674(89)90935-5. [DOI] [PubMed] [Google Scholar]
- Lech K., Anderson K., Brent R. DNA-bound Fos proteins activate transcription in yeast. Cell. 1988 Jan 29;52(2):179–184. doi: 10.1016/0092-8674(88)90506-5. [DOI] [PubMed] [Google Scholar]
- Lew D. J., Dulić V., Reed S. I. Isolation of three novel human cyclins by rescue of G1 cyclin (Cln) function in yeast. Cell. 1991 Sep 20;66(6):1197–1206. doi: 10.1016/0092-8674(91)90042-w. [DOI] [PubMed] [Google Scholar]
- Lüscher B., Eisenman R. N. New light on Myc and Myb. Part I. Myc. Genes Dev. 1990 Dec;4(12A):2025–2035. doi: 10.1101/gad.4.12a.2025. [DOI] [PubMed] [Google Scholar]
- Matsushime H., Ewen M. E., Strom D. K., Kato J. Y., Hanks S. K., Roussel M. F., Sherr C. J. Identification and properties of an atypical catalytic subunit (p34PSK-J3/cdk4) for mammalian D type G1 cyclins. Cell. 1992 Oct 16;71(2):323–334. doi: 10.1016/0092-8674(92)90360-o. [DOI] [PubMed] [Google Scholar]
- Matsushime H., Roussel M. F., Ashmun R. A., Sherr C. J. Colony-stimulating factor 1 regulates novel cyclins during the G1 phase of the cell cycle. Cell. 1991 May 17;65(4):701–713. doi: 10.1016/0092-8674(91)90101-4. [DOI] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989 Mar 10;56(5):777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Ohtsubo M., Roberts J. M. Cyclin-dependent regulation of G1 in mammalian fibroblasts. Science. 1993 Mar 26;259(5103):1908–1912. doi: 10.1126/science.8384376. [DOI] [PubMed] [Google Scholar]
- Pardee A. B. G1 events and regulation of cell proliferation. Science. 1989 Nov 3;246(4930):603–608. doi: 10.1126/science.2683075. [DOI] [PubMed] [Google Scholar]
- Penn L. J., Brooks M. W., Laufer E. M., Land H. Negative autoregulation of c-myc transcription. EMBO J. 1990 Apr;9(4):1113–1121. doi: 10.1002/j.1460-2075.1990.tb08217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak K., Kato J. Y., Solomon M. J., Sherr C. J., Massague J., Roberts J. M., Koff A. p27Kip1, a cyclin-Cdk inhibitor, links transforming growth factor-beta and contact inhibition to cell cycle arrest. Genes Dev. 1994 Jan;8(1):9–22. doi: 10.1101/gad.8.1.9. [DOI] [PubMed] [Google Scholar]
- Prendergast G. C., Cole M. D. Posttranscriptional regulation of cellular gene expression by the c-myc oncogene. Mol Cell Biol. 1989 Jan;9(1):124–134. doi: 10.1128/mcb.9.1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast G. C., Ziff E. B. Methylation-sensitive sequence-specific DNA binding by the c-Myc basic region. Science. 1991 Jan 11;251(4990):186–189. doi: 10.1126/science.1987636. [DOI] [PubMed] [Google Scholar]
- Prouty S. M., Hanson K. D., Boyle A. L., Brown J. R., Shichiri M., Follansbee M. R., Kang W., Sedivy J. M. A cell culture model system for genetic analyses of the cell cycle by targeted homologous recombination. Oncogene. 1993 Apr;8(4):899–907. [PubMed] [Google Scholar]
- Quelle D. E., Ashmun R. A., Shurtleff S. A., Kato J. Y., Bar-Sagi D., Roussel M. F., Sherr C. J. Overexpression of mouse D-type cyclins accelerates G1 phase in rodent fibroblasts. Genes Dev. 1993 Aug;7(8):1559–1571. doi: 10.1101/gad.7.8.1559. [DOI] [PubMed] [Google Scholar]
- Sherr C. J. Mammalian G1 cyclins. Cell. 1993 Jun 18;73(6):1059–1065. doi: 10.1016/0092-8674(93)90636-5. [DOI] [PubMed] [Google Scholar]
- Shichiri M., Hanson K. D., Sedivy J. M. Effects of c-myc expression on proliferation, quiescence, and the G0 to G1 transition in nontransformed cells. Cell Growth Differ. 1993 Feb;4(2):93–104. [PubMed] [Google Scholar]
- Spencer C. A., Groudine M. Control of c-myc regulation in normal and neoplastic cells. Adv Cancer Res. 1991;56:1–48. doi: 10.1016/s0065-230x(08)60476-5. [DOI] [PubMed] [Google Scholar]
- Stern D. F., Roberts A. B., Roche N. S., Sporn M. B., Weinberg R. A. Differential responsiveness of myc- and ras-transfected cells to growth factors: selective stimulation of myc-transfected cells by epidermal growth factor. Mol Cell Biol. 1986 Mar;6(3):870–877. doi: 10.1128/mcb.6.3.870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteeg R., Noordermeer I. A., Krüse-Wolters M., Ruiter D. J., Schrier P. I. c-myc down-regulates class I HLA expression in human melanomas. EMBO J. 1988 Apr;7(4):1023–1029. doi: 10.1002/j.1460-2075.1988.tb02909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters C. M., Littlewood T. D., Hancock D. C., Moore J. P., Evan G. I. c-myc protein expression in untransformed fibroblasts. Oncogene. 1991 May;6(5):797–805. [PubMed] [Google Scholar]
- Weintraub H., Davis R., Tapscott S., Thayer M., Krause M., Benezra R., Blackwell T. K., Turner D., Rupp R., Hollenberg S. The myoD gene family: nodal point during specification of the muscle cell lineage. Science. 1991 Feb 15;251(4995):761–766. doi: 10.1126/science.1846704. [DOI] [PubMed] [Google Scholar]
- Wickstrom E. L., Bacon T. A., Gonzalez A., Freeman D. L., Lyman G. H., Wickstrom E. Human promyelocytic leukemia HL-60 cell proliferation and c-myc protein expression are inhibited by an antisense pentadecadeoxynucleotide targeted against c-myc mRNA. Proc Natl Acad Sci U S A. 1988 Feb;85(4):1028–1032. doi: 10.1073/pnas.85.4.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B. S., Gilbert J. D., Freytag S. O. Overexpression of Myc suppresses CCAAT transcription factor/nuclear factor 1-dependent promoters in vivo. Mol Cell Biol. 1993 May;13(5):3093–3102. doi: 10.1128/mcb.13.5.3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel S., Harlow E. Distinct roles for cyclin-dependent kinases in cell cycle control. Science. 1993 Dec 24;262(5142):2050–2054. doi: 10.1126/science.8266103. [DOI] [PubMed] [Google Scholar]




