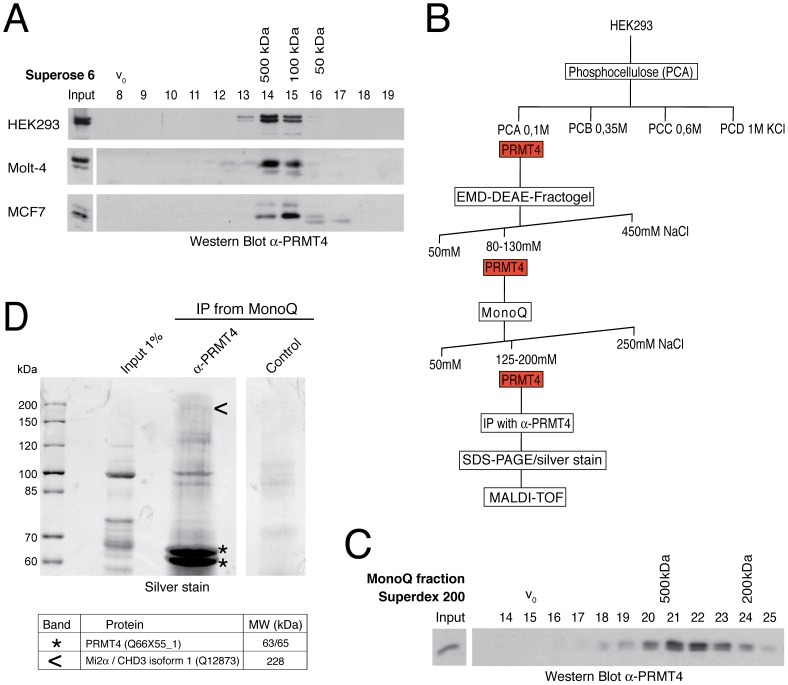
Figure 1. Identification of putative interaction partners of PRMT4.
A: PRMT4 resides within a high molecular weight protein complex. For size fractionation by gel filtration chromatography, whole-cell protein extracts were generated from HEK293, Molt-4 and MCF7 cells and subjected to Benzonase treatment. Protein extracts were applied to a Superose 6 column and 1 ml fractions were collected. Fractions (8–19) were stained by Western Blot analysis using anti-PRMT4 antibodies. The column was calibrated using standard protein markers. Accordingly, the size of the PRMT4-containing fractions and the void volume (V0) are indicated. B: Biochemical purification of endogenous PRMT4. Schematic representation of the chromatographic steps used to purify PRMT4-containing complexes from HEK293 protein extract. C: High molecular weight complexes of PRMT4 remain stably associated during purification procedure. PRMT4-containing MonoQ fractions were size-fractionated using a Superdex 200 column. After collection of fractions (500 µl each), fraction numbers 14–25 were stained by Western Blot analysis using anti-PRMT4 antibodies. The size of the PRMT4-containing fractions and the void volume (V0) are indicated. D: Affinity purification of endogenous PRMT4 by immunoprecipitation. PRMT4-containing MonoQ fractions were subjected to IP using polyclonal anti-PRMT4 antibodies compared to bead control. Input (1% = 12 µg) and immunoprecipitates were separated by SDS-PAGE. Silver-stained bands specifically detected in the anti-PRMT4 sample were excised and identified by MALDI-TOF peptide mass fingerprint analysis as PRMT4 (asterisks) and Mi2α (arrowheads). SDS-PAGE size markers (in kDa) are shown on the left. Two bands were identified as PRMT4, which are likely different isoforms (of 63 and 65 kDa), but which could not be distinguished by peptides in the mass spectrometry.