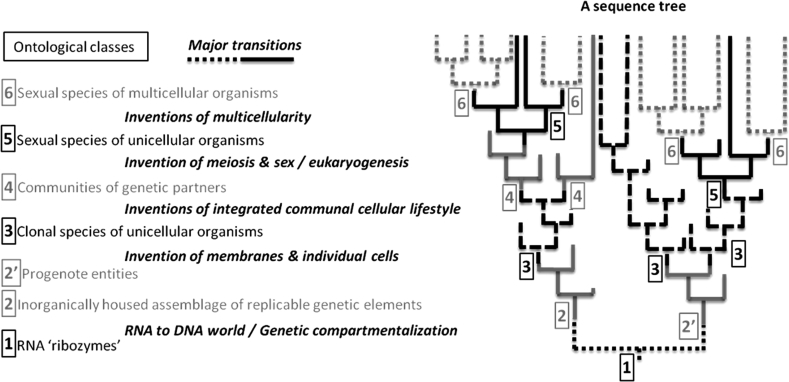
Fig. 1.
Decoupling of changes in sequence and carrier-ontology. The evolution of the relatively most stable entities used to track evolutionary changes in the microbial world (e.g. sequences from a well conserved gene family) can be represented by a tree. The branching and ancestral nodes in such a gene tree are commonly used to infer the presence of a particular type of evolutionary event (e.g. speciation, transfer, etc.), affecting a particular class of less stable entities (e.g. ancestral organism, species, etc.). However, the difference of time scales for the stabilisation of the most stable entities and that for the other entities that these slowly evolving sequences are supposed to be tracking makes it impossible to infer a direct correspondence between the changes recorded in the gene tree and those that have occurred in other entities. Dichotomies and divergences in the gene tree cannot be directly coupled to changes in the ontology of the gene carriers, and the evolutionary events affecting less stable entities of which this gene is a component. There is not necessarily information on the gene tree alone about what type of entities carried a particular ancestral gene form, and about whether and when these types of entities may have dramatically changed, i.e. when the evolution of novel classes of sequence-based entities resulted in major (hypothetical) evolutionary transitions. (Interested readers can see Gross and Bhattacharya (2010), Koonin and Martin (2005), and Woese (1990) for more details on some of these transitions.) Such decoupling means that gene trees are informative about gene history, but may not be informative about the evolution of other entities, contrary to what is assumed in a standard ontology, which couples the evolutionary history of parts and wholes. Fig. 1 represents important ontological transitions by changes in branch format. Because of these changes, relatively similar sequences in a branching lineage may reside in different types of (sometimes novel) entities with different time scales for their stabilisation. Interpreting a sequence tree with a single ontology from present to past will then provide a severely distorted view of evolution