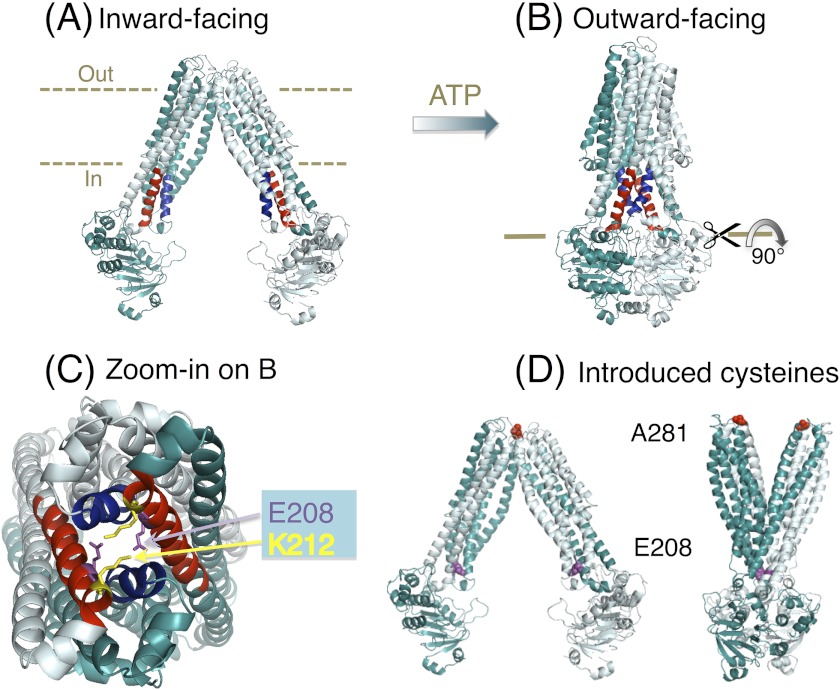
FIGURE 1.
MsbA structures and the tetrahelix bundle. A, nucleotide-free inward-facing structure of dimeric MsbA (Protein Data Bank code 3B5W, full model). B, AMP-PNP-bound outward-facing structure (Protein Data Bank code 3B60) (12). Residues Thr-121–Glu-133 in TM 3 and TM 3′ are in blue, and Asn-195–His-214 in TM 4 and TM 4′ are in red. C, zoomed in snapshot of the tetrahelix bundle viewed from the cytoplasmic side, after rotation of the MsbA dimer in B by 90°. Residues Ser-324 to Gln-582 in each monomer (containing the NBD) are not shown for clarity of presentation. Residues Glu-208 and Lys-212 (in the cytoplasmic extensions of TM 4 and TM 4′) are shown as purple and yellow sticks, respectively. D, residues Ala-281–Ala-281′ (red spheres) in dimeric MsbA are in close proximity in the nucleotide-free inward-facing conformation and distant in the AMP-PNP-bound outward-facing state, whereas Glu-208 to Glu-208′ residues (purple spheres) follow the opposite pattern. For further explanation, see under “A281C-A281C′ Cross-linking in Dimeric MsbA-cl Responds to Nucleotides.”