FIGURE 4.
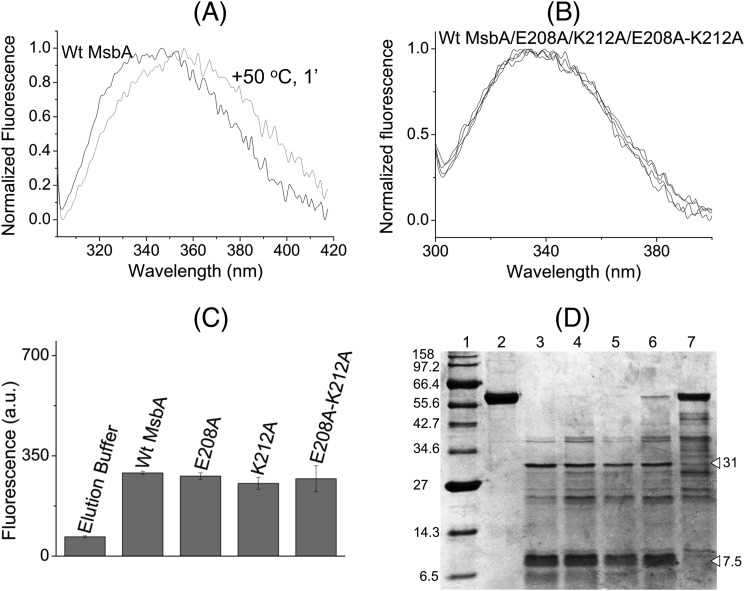
Alanine mutations in the tetrahelix bundle do not denature MsbA. WT MsbA and the mutant proteins showed similar overall protein folding that is different from heat-denatured MsbA, as deduced from intrinsic tryptophan fluorescence and trypsin digestion patterns. A, normalized spectral profiles of intrinsic tryptophan fluorescence measured for MsbA WT, without (black trace, labeled WT MsbA) or with (gray trace) denaturation for 1 min at 50 °C, revealed a denaturation-dependent right-shift in the emission maximum. B and C, alanine mutants E208A MsbA and K212A MsbA and the E208A/K212A double mutant gave overlapping tryptophan fluorescence traces (B) with similar peak fluorescence intensities at 328 nm as WT MsbA (n = 4) (C), and it did not show any signs of protein denaturation. Data are elution buffer-subtracted. Error bars represent mean ± S.E. D, trypsin digestions of purified MsbA proteins were examined on 12% SDS-PAGE (9 μg of protein/lane). Lane 1, molecular mass marker; lane 2, WT MsbA; lane 3, WT MsbA + trypsin; lane 4, E208A MsbA + trypsin; lane 5, K212A MsbA+ trypsin; lane 6, E208A/K212A MsbA + trypsin; lane 7, WT MsbA (denatured for 1 min at 50 °C) + trypsin. White triangles refer to major signals for WT MsbA (31 and 7.5 kDa) that were absent for heat-denatured MsbA. Band patterns are typical for observations in four independent experiments.