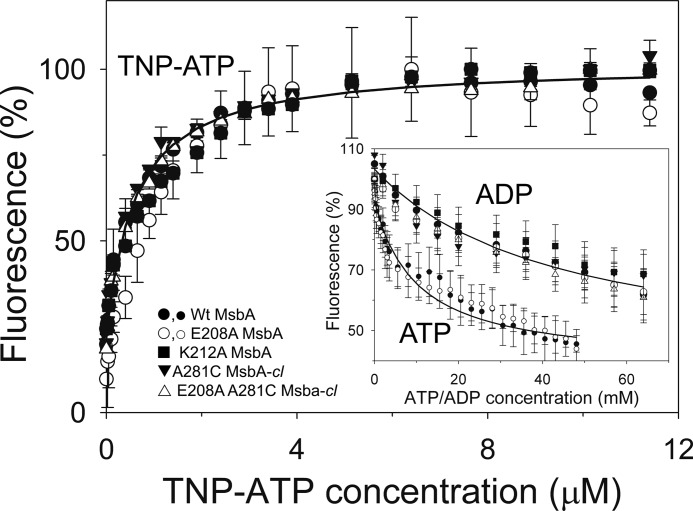
FIGURE 5.
MsbA mutants used in this study exhibit WT-like binding affinities for nucleotides. TNP-ATP binding and ADP-dependent and ATP-dependent TNP-ATP displacement (inset) were determined in triplicate (n = 3) using purified MsbA proteins in detergent solution (large symbols for TNP-ATP binding and its displacement by ADP; small open and closed circles for displacement of TNP-ATP binding by ATP). ATP concentration scale is ×50 the actual ATP concentration. The IC50 values obtained in the TNP-ATP displacement experiments were used in Equation 3 under “Binding Assays” to calculate the dissociation constants for binding of ADP (KdADP) and ATP (KdATP). Values for KdADP, KdATP, and the dissociation constant for binding of TNP-ATP (KdTNP-ATP) are listed in the text and in Table 1. Error bars represent mean ± S.E.