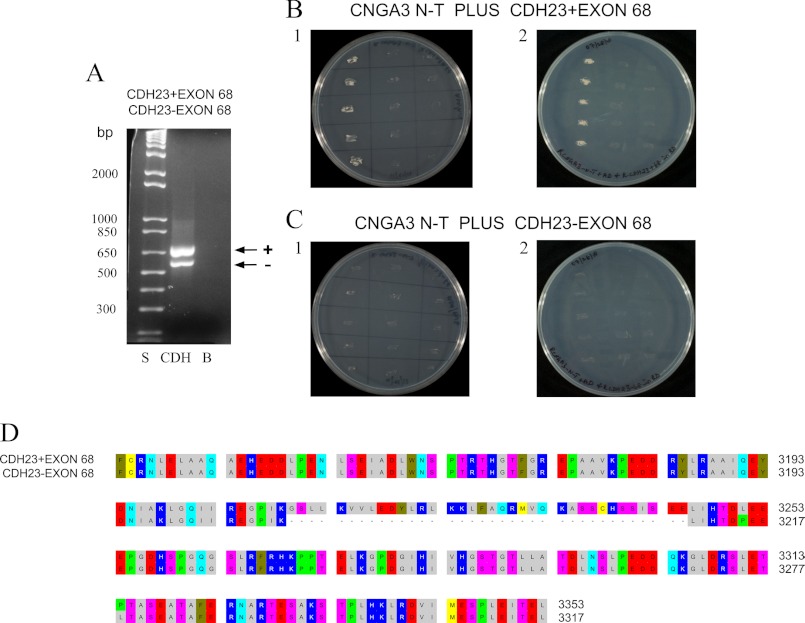
FIGURE 2.
Interaction of CNGA3 with CDH23 +68 and −68 in yeast two-hybrid mating protocols. A, CDH23 +68 and CDH23 −68 messages were expressed in rat organ of Corti. PCR amplification is shown of 660-bp product for CDH23 +68 (lane CDH, arrow +) and 552-bp for CDH23 −68 (lane CDH, arrow −) with lane S = 1 kb plus standards ladder (Invitrogen) and lane B = negative control for which cDNA was omitted. B1, yeast two-hybrid co-transformation assays with replicate streakings show CNGA3 binding to CDH23 +68. First lane, CNGA3 amino terminus (CNGA3 N-T) in bait vector and CDH23 +68 in prey vector; second lane, CNGA3 bait vector and empty prey vector as negative control; third lane, empty bait vector and CDH23 +68 in prey vector as another negative control. B2, shown is reversal of bait and prey sequence for binding of CNGA3 to CDH23 +68. First lane, CNGA3 in prey vector and CDH23 +68 in bait vector; second lane, CNGA3 in prey vector and empty bait vector as negative control; third lane, CDH23 +68 in bait vector and empty prey vector as another negative control. C1, CNGA3 does not bind to CDH23 −68 in yeast two-hybrid co-transformation assays. First lane, CNGA3 in bait vector and CDH23 −68 in prey vector; second lane, CNGA3 in bait vector and empty prey vector as negative control; third lane, empty bait vector and CDH23 −68 in prey vector as another negative control. C2, reversal of bait and prey sequence for CNGA3 and CDH23 −68 is shown. First lane, CNGA3 in prey vector and CDH23 −68 in bait vector; second lane, CNGA3 in prey vector and empty bait vector as negative control; third lane, CDH23 −68 in bait vector and empty prey vector as another negative control. D, shown is alignment (with OMIGA 2.0) of cytoplasmic carboxyl domain of rat CDH23 with amino acid numbering according to NP_446096.1. Alternative splicing generates CDH23 +68 (EDL93044) and CDH23 −68 (NP_446096.1) that differ in their cytoplasmic domain by inclusion/exclusion of exon 68. These sequences were used as prey/bait for binding to CNGA3.