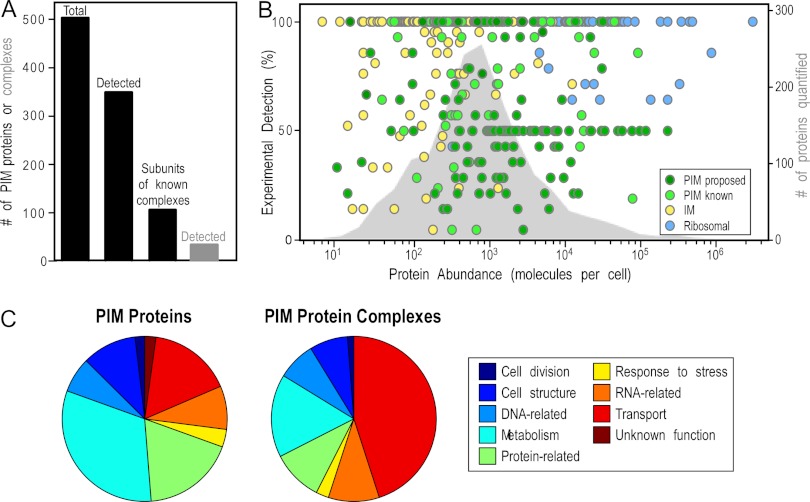
Fig. 3.
Features of the peripherome. A, combined protein identifications of PIM and protein complexes from two approaches. The proposed total E. coli peripherome is composed of at least 503 PIM proteins (47% from membrane proteolysis, 16% from complexomics, and 37% from the literature and re-annotation). In this study, a total of 342 peripheral proteins were identified by our approaches; 106 of them are known to participate in 63 PIM complexes. A total of 34 heteromultimeric complexes were identified experimentally. B, abundance of PIM proteins. The abundance of the experimentally detected proteins (colored circles, supplemental Table S1A, Fig. 4) is depicted using quantitation values previously determined for E. coli K-12. The right-hand y-axis represents the percentage of detection and expresses the number of times a protein has been identified relative to the number of experimental repeats. Peripheral proteins are shown with green circles, and ribosomal and integral membrane proteins are denoted by blue and yellow circles, respectively. The abundance distribution of 1783 proteins available in the literature is plotted on the background (gray area) (33–35). C, functional assignment of PIM proteins. Distribution of PIM proteins based on the MultiFun cell function classification system (87). Gene ontology information of individual proteins was collected from EcoCyc and Uniprot (3, 5). Most of the PIM proteins are involved in metabolism (i.e. energy metabolism, electron transport, amino acid biosynthesis) and in information transfer (protein-, DNA-, RNA-related) processes of the cell (supplemental Table S7).