Fig. 4.
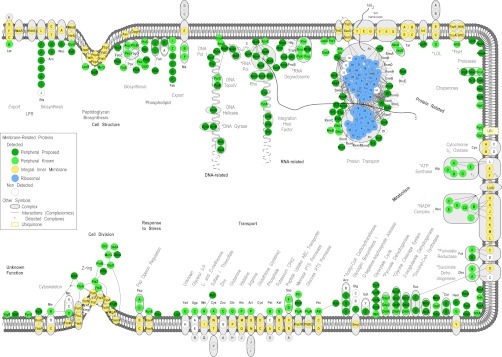
Bird's-eye view of the E. coli peripherome. A schematic longitudinal section of an E. coli dividing cell is shown. A section of the inner membrane is depicted; the rest of the cell envelope is omitted for simplicity. Based on MultiFun classification, PIM proteins (in green circles) participate in nine cellular processes (indicated with bold letters): cell structure; DNA-, RNA-, and protein related; metabolism; transport; response to stress; and cell division. Sub-categories of these processes are indicated with bold gray letters. The proteins are grouped according to known complexes (encircled in gray color schemes) and pathways, together with their integral inner membrane partners (yellow circles) where known (3). Protein–protein interactions derived from tandem-affinity purification experiments (88) are also mapped (where available) and depicted with connecting lines. For visualization purposes, the protein–protein interactions were integrated and loaded in Cytoscape (3). The interactions shown are not necessarily simultaneous and might have derived from the capture of different holoenzyme variants with different subunits. Proteins drawn in the peptidoglycan and phospholipid biosynthesis and cell division are based on known interactions and were downloaded only from IntAct (88). Proteins of the ribosome, arranged in the 50S and the 30S subunits, are drawn in the membrane's vicinity (blue circles). Following translation in the ribosome, proteins (indicated with a thick gray scribbled line) are recognized co- or post-translationally by the ribosome-bound signal recognition particle or the SecA and/or SecB chaperones, respectively, and are destined for the membrane for secretion through the Sec translocase (89). A number of proteins, previously annotated as cytoplasmic, are shown to associate with the inner membrane and are proposed as new PIM proteins. These are represented with dark green circles, whereas previously known and curated (in this study) peripheral proteins are featured with light green circles. For a comprehensive view of the mapped complexes, some periplasmic proteins are also drawn (where available) on the membrane outer surface. Complexes identified are indicated with a red star. Proteins with no functional annotations are indicated as “Unknown Function.” For the complete list, annotation, and description of protein complexes identified, see supplemental Table S4.
