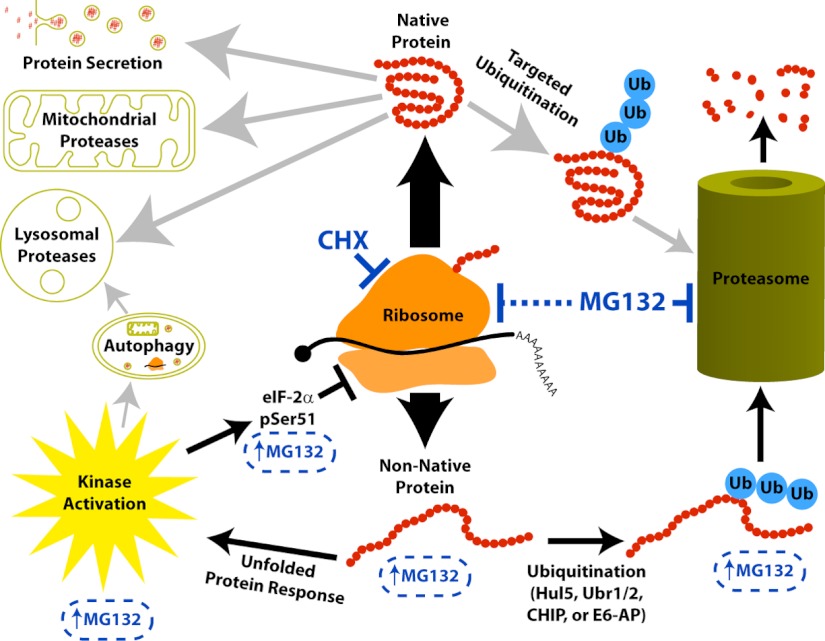
Fig. 6.
Model for proteome-wide responses after translation inhibition or proteasome inhibition. The ribosome produces both native protein and nonnative protein. Native protein may be degraded by mechanisms that may differ depending on subcellular localization (indicated by the gray arrows). Cycloheximide treatment allows us to observe this depletion as a decreased abundance of a protein over time. Under normal conditions the nonnative protein is quickly ubiquitinated and degraded by the proteasome. Proteasome inhibition by MG132 treatment blocks the degradation of both native proteins and nonnative proteins. This induces a build-up of nonnative protein that causes an unfolded protein response. One outcome of this response to MG132 is the inhibition of protein synthesis by eIF-2α pSer51 phosphorylation. Therefore, because MG132 simultaneously blocks the proteasome and protein synthesis, we observe very few proteins increasing in abundance after proteasome inhibition and only the subset of RDPs depleted by nonproteasomal mechanisms decrease in abundance.