Figure 2.
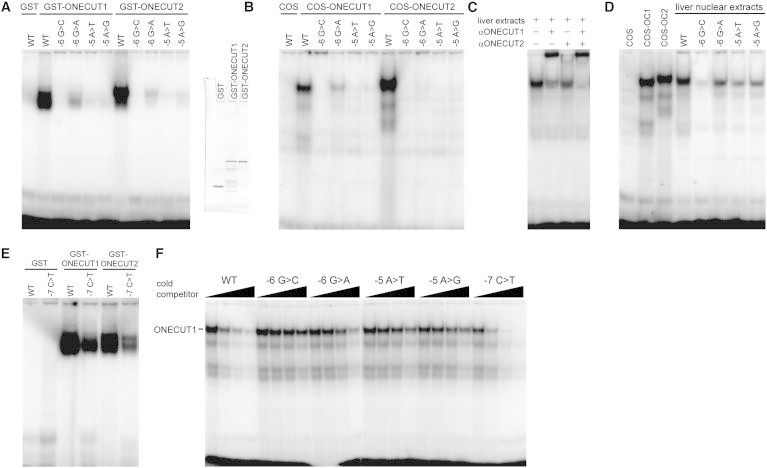
ONECUT Proteins Recognize the −5/−6 Region of the Human F9 Promoter, and Binding Is Disrupted by Leyden Mutations
(A–F) EMSAs using wild-type (WT) and mutant probes spanning the −5/−6 region.
(A) ONECUT1 and ONECUT2 GST-fusion proteins purified from bacteria. Coomassie staining is shown on the right to indicate the level and purity of the recombinant proteins used.
(B) Nuclear extracts from COS cells expressing full-length ONECUT1 and ONECUT2.
(C) Murine liver nuclear extracts with antibodies against ONECUT1 and ONECUT2.
(D) Murine liver nuclear extracts with WT and mutant probes. Nuclear extracts from COS cells transfected with ONECUT1 and ONECUT2 were included as positive controls.
(E) ONECUT1 and ONECUT2 GST-fusion proteins purified from bacteria. Equivalent levels of protein were added as in (A).
(F) A competition assay using radiolabeled WT probe (1.6 ng per lane) and cold competitor probes (0 ng, 16 ng, 160 ng, and 1.6 μg, that is, 0×, 10×, 100×, and 1,000× excesses, respectively) compares the ability of the different mutants to compete for binding by ONECUT1. Two microliters of nuclear extracts from COS cells expressing ONECUT1 was used per lane. Murine liver tissue was obtained under ethics approval number 09/128A (Animal Care and Ethics Committee, University of New South Wales). The following protein abbreviations are used: OC1, ONECUT1; and OC2, ONECUT2.
