Figure 3.
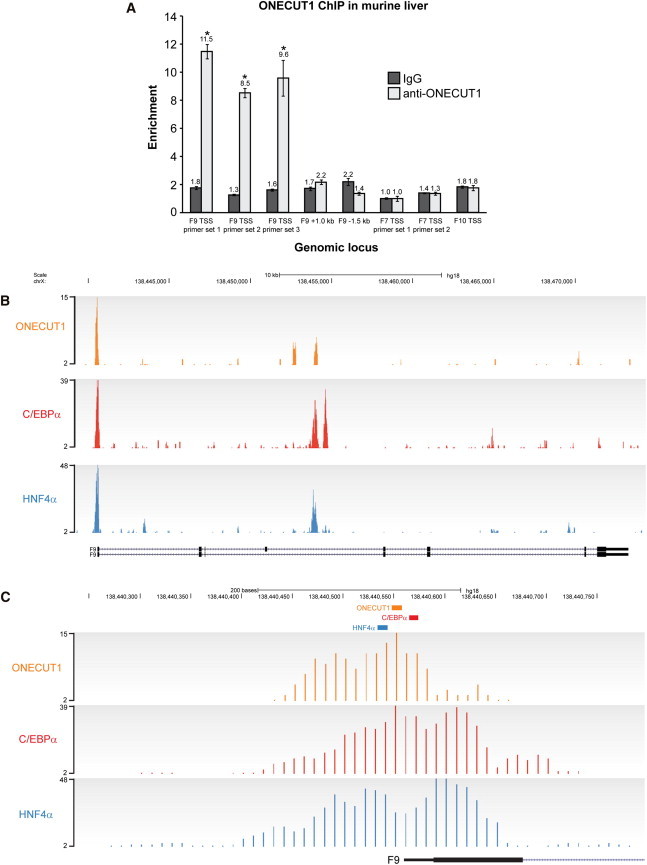
ONECUT1 Binds the F9 Promoter In Vivo along with HNF4α and C/EBPα
(A) ONECUT1 ChIP using mouse liver tissue. Primer sets are specific to the TSS of F9 and to 1.0 kb downstream and 1.5 kb upstream. Negative-control primers specific to the promoter regions of F7 and F10 were included. Enrichment was normalized to input, and the lowest values for immunoglobulin G (IgG) and anti-ONECUT1 were set to 1.0. Error bars represent the SEM, and n = 3 for each data point. ∗p < 0.05 (two-tailed t test) compared to the anti-ONECUT1 enrichment at each of the F9 distal regions and the F7 and F10 promoters.
(B–C) Human-liver-tissue ChIP-Seq tracks for ONECUT1, C/EBPα, and HNF4α across the F9 locus (B) and around the TSS (C).
(C) The HNF4α, ONECUT1, and C/EBPα binding sites at nucleotides −20, −5/−6, and +10, respectively, are denoted by colored boxes. Murine liver tissue was obtained under ethics approval number 09/128A (Animal Care and Ethics Committee, University of New South Wales). ChIP sequencing reads and input samples were aligned with MAQ with default parameters to NCBI Genome browser build 36. All sequence, genome-annotation, and comparative-genomics data were taken from Ensembl release 52. After alignment, binding events were discovered with a dynamic programming algorithm (SWEMBL) with the parameter “-R 0.005 -i –S” as previously described.27
