Abstract
“Candidatus Portiera aleyrodidarum” is the primary endosymbiont of whiteflies. We report two complete genome sequences of this bacterium from the worldwide invasive B and Q biotypes of the whitefly Bemisia tabaci. Differences in the two genome sequences may add insights into the complex differences in the biology of both biotypes.
TEXT
Obligate endosymbionts living inside their hosts have repeatedly evolved in several lineages of insects from free-living bacterial ancestors and maintained this relationship for millions of years (1). During this long-lasting symbiosis, insect hosts have provided shelter for their obligate endosymbionts and, in return, these endosymbionts have evolved metabolic pathways for the synthesis of essential nutrients for their hosts, nutrients lacking in the insects' natural diet, thus ensuring the species' persistence. Because of the transition from free-living to symbiotic bacteria, recent genome sequencing projects have shown several extremely striking patterns of genome evolution in several lineages of obligate endosymbionts (2). These patterns include extensive genome erosion leading to the smallest cellular genomes (<1 Mb), elevated rates of nonsynonymous substitutions, depleted mobile elements and repeated sequences, and extreme genome stasis lasting millions of years (1, 2).
“Candidatus Portiera aleyrodidarum” (here referred to as “Ca. Portiera”) is the obligate primary endosymbiotic bacterium hosted by whiteflies, including the sweet potato whitefly Bemisia tabaci (3) (Fig. 1). B. tabaci is one of the most globally damaging agricultural pests, causing annual losses estimated at $1 to 2 billion and is rated one of the top 100 invasive species worldwide (4, 5). Similar to other obligate bacteria living in sap-sucking insects, “Ca. Portiera” is thought to provide essential nutrients to whiteflies and this relationship has been maintained for approximately 180 million years.
Fig 1.
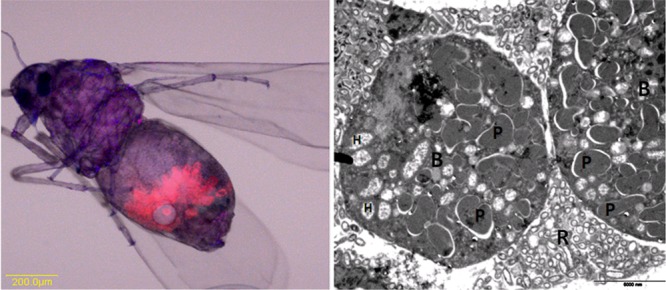
(Left panel) “Ca. Portiera” localization in adult female B. tabaci by fluorescence in situ hybridization analysis with a probe specific for “Ca. Portiera” (red). Note that “Ca. Portiera” is restricted to bacteriocyte cells in the insect abdomen. (Right panel) transmission electron microscopic image showing two bacteriocytes (B) full of “Ca. Portiera” (P) and some cells of the secondary endosymbiont Hamiltonella (H) and surrounded by the secondary symbiont Rickettsia (R), which is scattered in the hemolymph. The bar in the right panel is 5 μm.
B. tabaci is a species complex composed of at least 24 morphologically indistinguishable species (6, 7). The most predominant and damaging biotypes are B and Q, which differ considerably with regard to various fitness parameters. Here we report the sequences, assembly, and comparison of “Ca. Portiera” genomes from these two biotypes. One biotype B whitefly strain, named B-HRs, and one biotype Q whitefly strain, named Q-AWRs, were used for DNA extraction, sequencing, and assembly as described in the supplemental material.
General assembly and coding of “Ca. Portiera” genomes.
All assembled contigs from the B and Q strains were blasted against the bacterial genome database and the 40-kb “Ca. Portiera” sequences deposited in the GenBank database (blastn; E, <1e-20). The “Ca. Portiera” genome assembly is a 351-kb circular molecule that is highly AT biased (73%) and larger than those of other previously reported insect primary endosymbionts such as Tremblaya from mealybugs with a 139-kb genome size (8), Hodgkinia from cicadas with a 144-kb genome size (9), and Carsonella from psyllids with a 160-kb genome size (10) but smaller than those of other primary endosymbionts such as Buchnera from aphids with a 640-kb genome size (11), Blattabacterium from cockroaches with a 630-kb genome size (12), and Wigglesworthia from tsetse flies with a 698-kb genome size (13). The median sequence coverages are 223- and 185-fold for the “Ca. Portiera” genomes from the B biotype (here referred to as “Ca. Portiera” WB) and the Q biotype (here referred to as “Ca. Portiera” WQ), respectively. Not surprisingly, “Ca. Portiera” WB and WQ share high similarity in alignable regions (99.8%) and have the same gene order (Fig. 2), except that the “Ca. Portiera” WQ genome is 700 bp shorter than that of “Ca. Portiera” WB.
Fig 2.
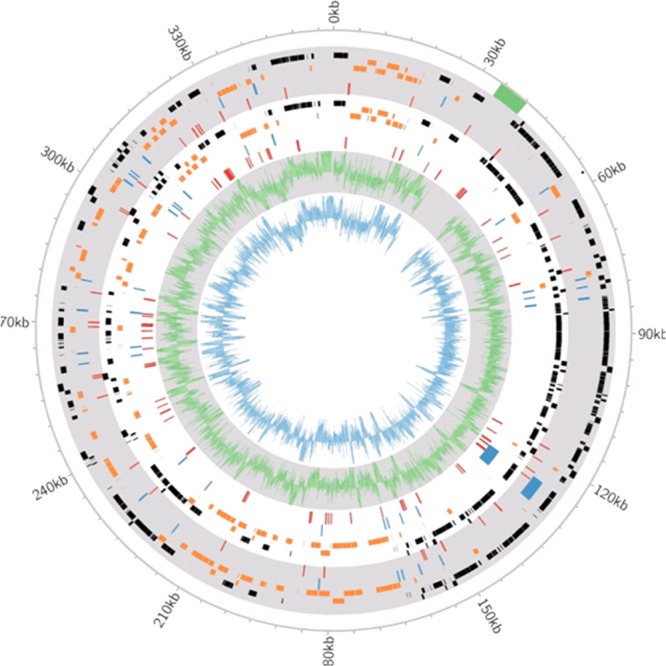
“Ca. Portiera” genome. From outside to inside, the tracks depict (i) features of “Ca. Portiera”_WQ (gray), (ii) features of “Ca. Portiera”_WB (white), (iii) the GC content of “Ca. Portiera”_WQ in a 50-bp window (green histogram on a gray background), and (iv) the GC content of “Ca. Portiera”_WB in a 50-bp window (blue histogram on a white background). The bars in black are genes on the plus strands of the genomes, the bars in orange are genes on the minus strands of the genomes, the bars in blue are the positions of RNA (tRNA and rRNA)-encoding genes, and the red bars are the regions of each genome that differ from the reference genome (see Materials and Methods). The labels at the tick marks refer to NC_018507.1. The bar in green is the 6-kb region that differs between our assemblies and the reference genome.
Unlike other endosymbionts that have highly AT-biased genomes and higher coding densities (e.g., Buchnera aphidicola [87.7%] and Carsonella ruddii [97.3%]), the percentages of the “Ca. Portiera” genomes that are made up of protein-coding genes are strikingly low (69.0% for “Ca. Portiera” WB and 70.3% for “Ca. Portiera” WQ). Both “Ca. Portiera” genomes encode 3 rRNAs and 33 tRNAs. “Ca. Portiera” WB has 271 protein-coding genes, and “Ca. Portiera” WQ has 278 protein-coding genes (see Tables S1a, S1b, and S1c in the supplemental material). Similar to other obligate endosymbionts from sap-feeding insects whose genomes have been sequenced, enrichment for genes involved in essential amino acid biosynthesis was observed, while genes involved in processes such as membrane transport, cell wall/capsule, motility, DNA repair, regulation, and cell signaling were not found (see Fig. S1 in the supplemental material). These results support the hypothesis that primary endosymbionts primarily supply their hosts with amino acids. Additionally, three carotenoid biosynthesis genes were identified in the two “Ca. Portiera” genomes, similar to the recently observed carotenoid genes in “Ca. Portiera” from additional biotype B strains (14).
Low polymorphism rate in the “Ca. Portiera” genome.
As our DNA sample was pooled from over 100 individuals and there are thousands of copies of “Ca. Portiera” bacteria in each individual, we mapped all of the reads to the final assembly to estimate the allele frequency of possible polymorphic sites. To minimize the possibility of PCR-mediated sequencing errors and remove rare alleles, alleles with at least three copies were considered (after the removal of PCR replicates by using SAMtools) (15). We found that there are only 301 polymorphic sites (<0.1%; minor allele, at least three copies; see Table S2 in the supplemental material) in “Ca. Portiera” WQ, and similar results were obtained with “Ca. Portiera” WB (809 polymorphic sites; 0.2%). Overall, the low single-nucleotide polymorphism rate can be attributed to the pooled DNA samples that originated from highly inbred whitefly strains.
Divergence between B. tabaci B and Q “Ca. Portiera” genomes.
By mapping all of the reads from “Ca. Portiera” WQ to “Ca. Portiera” WB, we found 71 regions (varying in size from 2 to 266 bp; total, 3,963 bp) not present in the “Ca. Portiera” WQ genome. Of these 71 regions, 8 are located within genic regions, 11 are near the 5′- or 3′-end region, and 51 are intergenic (see Tables S3 and S4 in the supplemental material). Unsurprisingly, most of the unique regions (51/71; 71.8%) are located within intergenic regions. By mapping all of the reads from the B-HRs strain to the “Ca. Portiera” WQ genome assembly, we found 38 regions (varying in size from 2 to 180 bp; total, 1,923 bp) not present in the “Ca. Portiera” WB genome (see Table S3 in the supplemental material). Of these 38 regions, 7 are located within genic regions, 8 are near the 5′- or 3′-end region, and 23 are intergenic (see Tables S3 and S5 in the supplemental material). Interestingly, there is an 8-bp unique region only 30 bp upstream of the gene for an ABC transporter and ATP-binding protein, a 69-bp unique region upstream of the gene for transfer mRNA-binding protein SmpB, and a 13-bp region 20 bp downstream of the gene for a glutaredoxin-related protein. These short unique sequences may impact the efficiency of transcription of these genes.
About two-thirds of the unique regions identified in each “Ca. Portiera” genome are located in intergenic regions and are more likely to be neutral with respect to the fitness of “Ca. Portiera.” For the remaining one-third of the unique regions located within or near the protein-coding gene regions, they are more likely to have some functional roles in the fitness of “Ca. Portiera.” Most of those regions encode short hypothetical proteins with undiscovered functions.
A 6-kb region that differs between strains.
During the preparation of this report, a complete “Ca. Portiera” genome sequence from a biotype B whitefly collected in the United States was released (14) (here referred to as “Ca. Portiera” B-BT). Comparison of the “Ca. Portiera” WB genome assembly we obtained in our study with the recently released B-BT genome assembly revealed that the two are similar except for a 6-kb region (positions 34181 to 40300) that does not exist in our assembly. This region contains three genes (yidC, membrane protein insertase; trmE, GTP-binding protein; gidA, tRNA uridine 5-carboxymethylaminomethyl modification enzyme). By mapping all of our reads to the “Ca. Portiera” B-BT genome assembly, we found that there was no single read linking this region to the rest of the genome, while the rest of the sequences mapped with at least 3-fold coverage. Interestingly, we found this 6-kb region as a separate self-circled contig in our initial assemblies in both biotypes B and Q. In the B biotype, its coverage is 98-fold, which is half coverage of the “Ca. Portiera” WB genome. In the Q biotype, its coverage is 176-fold, which is similar to the coverage of the “Ca. Portiera” WQ genome. The difference of this 6-kb region between our assembly and the “Ca. Portiera” B-BT genome assembly could be due to the different natures of the strains used. There is some evidence, including a large number of reads and PCR amplification of a flanking region, that supports the existence of two conformations in the whitefly B-BT strain that was recently released (14).
Nucleotide sequence accession numbers.
The complete chromosome sequences determined in this study have been deposited in the GenBank database (“Ca. Portiera” WQ, accession no. CP003867; “Ca. Portiera” WB, accession no. CP003868).
Supplementary Material
ACKNOWLEDGMENTS
This research was partially supported by grant IS-4062-07 from the United States-Israel Binational Agricultural Research and Development Fund (BARD) and by research grant 887/07 from the Israel Science Foundation to M.G. We thank the Chicago Center for Systems Biology for the Research Experiences for Undergraduates (REU; NIH P50 GM081892) fellowship offered to K.P.W.
Footnotes
Published ahead of print 11 January 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02976-12.
REFERENCES
- 1. McCutcheon JP, Moran NA. 2012. Extreme genome reduction in symbiotic bacteria. Nat. Rev. Microbiol. 10:13–26 [DOI] [PubMed] [Google Scholar]
- 2. Wernegreen JJ. 2002. Genome evolution in bacterial endosymbionts of insects. Nat. Rev. Genet. 3:850–861 [DOI] [PubMed] [Google Scholar]
- 3. Baumann P. 2005. Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu. Rev. Microbiol. 59:155–189 [DOI] [PubMed] [Google Scholar]
- 4. Hu J, De Barro P, Zhao H, Wang J, Nardi F, Liu SS. 2011. An extensive field survey combined with a phylogenetic analysis reveals rapid and widespread invasion of two alien whiteflies in China. PLoS One 6:e16061 doi:10.1371/journal.pone.0016061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kontsedalova S, Abu-Mocha F, Lebedeva G, Czosnekb H, Horowitzc AR, Ghanim M. 2012. Bemisia tabaci biotype dynamics and resistance to insecticides in Israel during the years 2008-2010. J. Integr. Agric. 11:312–320 [Google Scholar]
- 6. Alemandri V, De Barro P, Bejerman N, Arguello Caro EB, Dumon AD, Mattio MF, Rodriguez SM, Truoli G. 2012. Species within the Bemisia tabaci (Hemiptera: Aleyrodidae) complex in soybean and bean crops in Argentina. J. Econ. Entomol. 105:48–53 [DOI] [PubMed] [Google Scholar]
- 7. De Barro PJ. 2005. Genetic structure of the whitefly Bemisia tabaci in the Asia-Pacific region revealed using microsatellite markers. Mol. Ecol. 14:3695–3718 [DOI] [PubMed] [Google Scholar]
- 8. López-Madrigal S, Latorre A, Porcar M, Moya A, Gil R. 2011. Complete genome sequence of “Candidatus Tremblaya princeps” strain PCVAL, an intriguing translational machine below the living-cell status. J. Bacteriol. 193:5587–5588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McCutcheon JP, McDonald BR, Moran NA. 2009. Origin of an alternative genetic code in the extremely small and GC-rich genome of a bacterial symbiont. PLoS Genet. 5:e1000565 doi:10.1371/journal.pgen.1000565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nakabachi A, Yamashita A, Toh H, Ishikawa H, Dunbar HE, Moran NA, Hattori M. 2006. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science 314:267 doi:10.1126/science.1134196 [DOI] [PubMed] [Google Scholar]
- 11. Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407:81–86 [DOI] [PubMed] [Google Scholar]
- 12. López-Sánchez MJ, Neef A, Patiño-Navarrete R, Navarro L, Jiménez R, Latorre A, Moya A. 2008. Blattabacteria, the endosymbionts of cockroaches, have small genome sizes and high genome copy numbers. Environ. Microbiol. 10:3417–3422 [DOI] [PubMed] [Google Scholar]
- 13. Akman L, Yamashita A, Watanabe H, Oshima K, Shiba T, Hattori M, Aksoy S. 2002. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat. Genet. 32:402–407 [DOI] [PubMed] [Google Scholar]
- 14. Sloan DB, Moran NA. 2012. Endosymbiotic bacteria as a source of carotenoids in whiteflies. Biol. Lett. 8:986–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup 2009. The sequence alignment/map (SAM) format and SAMtools. Bioinformatics 25:2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


