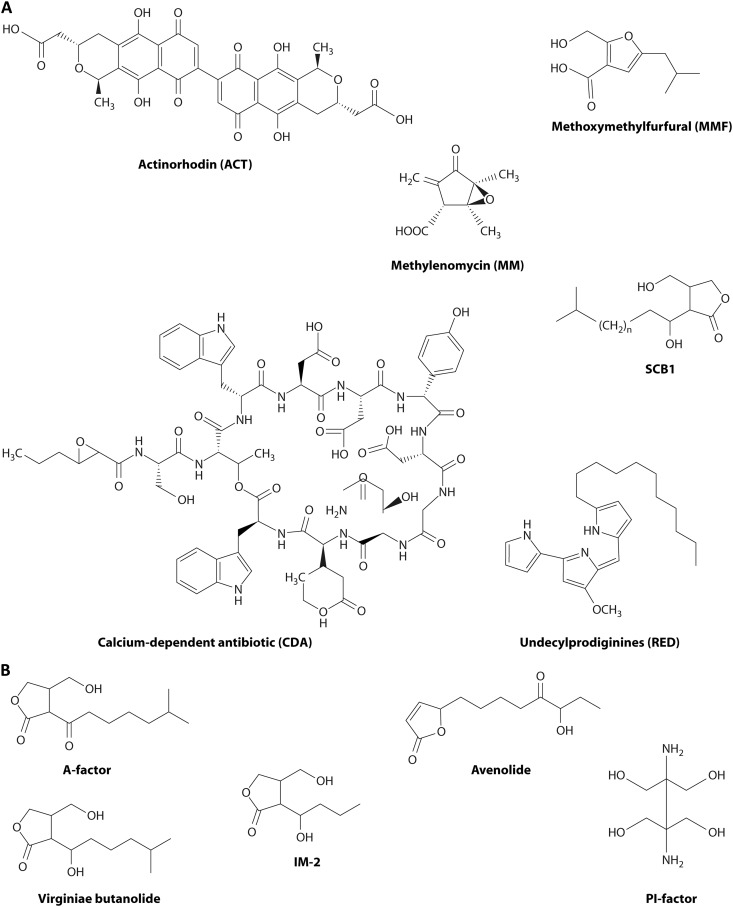
Fig 1.
Diverse antibiotics and autoregulator molecules produced by Streptomyces coelicolor A3(2) and some other streptomycetes. (A) The compounds from S. coelicolor. Actinorhodin (ACT) is a red/blue pH-indicating benzoisochromanequinone made by a type II polyketide synthase-based pathway involving a 22-gene cluster (291). Undecylprodiginines (REDs) are red hydrophobic tripyrroles made by a fatty acid synthase-like pathway involving a 22-gene cluster (292). Methylenomycin A (MM) is an epoxycyclopentenone made by an unusual pathway encoded by 11 genes located on the linear plasmid SCP1 (57). The calcium-dependent antibiotic (CDA) is a lipopeptide made by a route involving nonribosomal peptide synthases and specified by a 48-gene cluster (293). The autoregulator SCB1 (240) is one of several gamma-butyrolactone congeners whose synthesis involves three genes linked to the 19-gene cluster for biosynthesis of a structurally incompletely characterized yellow antibiotic (S. coelicolor polyketide [CPK]) (41, 42, 294). Methylenomycin furans (MMFs) are autoregulators that control MM biosynthesis, and they are made by a pathway resembling that of gamma-butyrolactones that involves three biosynthetic and two regulatory genes next to the MM biosynthetic cluster (58). (B) Autoregulatory molecules from other streptomycetes. A-factor regulates streptomycin biosynthesis in S. griseus (295). Virginiae butanolides (VBs) induce the coordinated production of virginiamycin M and virginiamycin S, synergistically acting but biosynthetically distinct antibiotics in S. virginiae (235). IM-2 controls the production of showdomycin and minimycin in S. lavendulae (237). PI factor enhances pimaricin production in S. natalensis (239). Avenolide is required at nanomolar concentrations for the onset of avermectin biosynthesis in S. avermitilis (238) (avenolide homologs are also found in S. fradiae, Streptomyces ghanaensis, and S. griseoauranticus).