Table 1.
Genes or operons in Escherichia coli known or predicted to be regulated with a DNA loop responsible for transcriptional repressiona
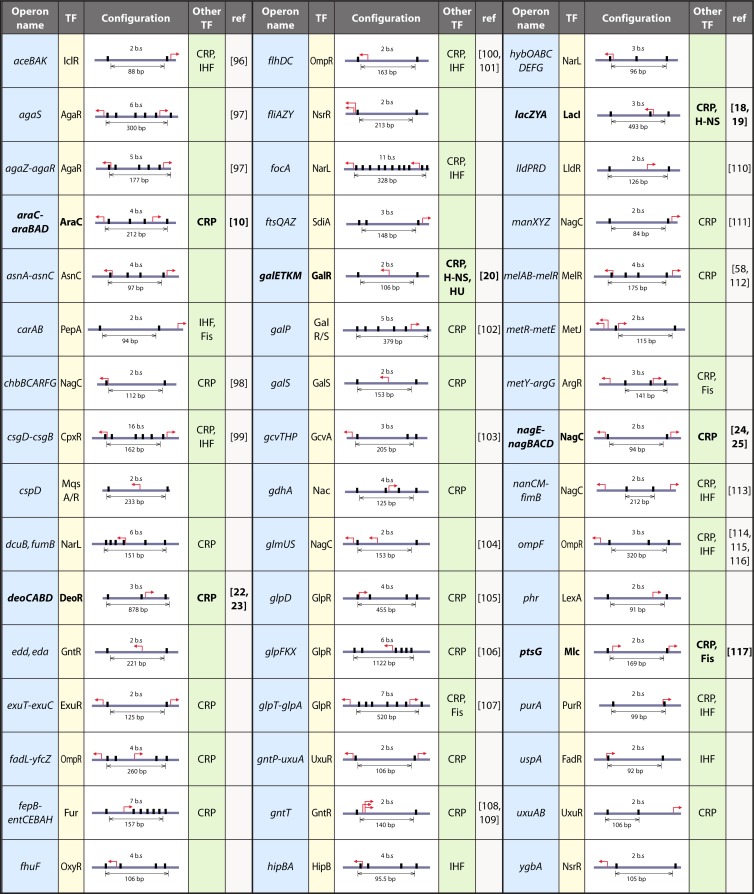
Data under the column heads are as follows. TF, transcription factor forming or predicted to form the DNA loop. Configuration, configuration of the binding sites responsible for DNA loop formation: the black boxes show the positions of the repressor binding sites relative to the transcription start site, indicated by a bent arrow, and the total number of binding sites (bs) and the distance between the first and last sites are indicated. Other TF, includes other transcription factors (CRP, H-NS, IHF, Fis, or HU) binding in the regulatory region that could have an impact on DNA loop formation. ref, reference(s) to the literature that validate or suggest regulation involving a DNA loop mechanism for this operon or give evidence against looping. Boldface indicates those operons where the loop has been clearly demonstrated.
