Fig 1.
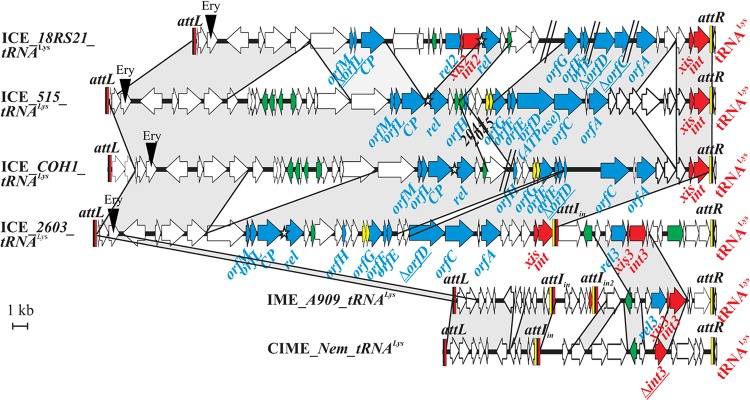
Open reading frame (ORF) organization and comparison of the elements integrated into the 3′ end of the tRNALys CTT gene in S. agalactiae 18RS21, 515, COH1, 2603V/R, A909, and Nem316. ORFs appear as arrows (truncated genes are indicated by a delta letter and underlined). The name of the gene where the element is integrated appears in red. Genes of the conjugation module are indicated with blue arrows, genes of the regulation module are indicated with green arrows, and genes of the recombination module are indicated with red arrows. When a putative domain has been found for the gene product or a putative function can be assigned to its product, the gene is named accordingly (rel for relaxase, CP for coupling protein, ATPase, xis for excisionase, and int for integrase). Other genes were named according to their similarity to ICESt1/St3 of S. thermophilus. The different genes encoding integrase, excisionase, and relaxase have been given different numbers. The putative toxin-antitoxin system (SAL2044-2045 and homologs in the other ICEs) appear as a yellow ORFs. The putative oriT is indicated by a star. Recombination sites are drawn as vertical rectangles. Black rectangles indicate identical sequences found in attL, attR, and attI sites. Yellow rectangles indicate the arm of attR sites and the related arm of attI sites, and red rectangles indicate the arm of attL sites and the related arms of attI sites. Protein identity higher than 80% is indicated in gray. Gaps in the genome due to missing contigs are indicated by a double slash.
