Fig 1.
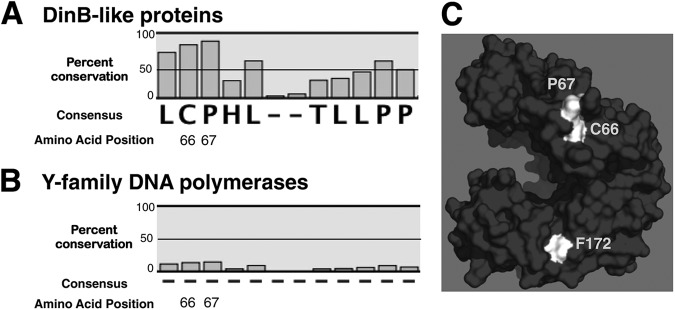
C66 and P67 are highly conserved noncatalytic residues in DinB-like proteins only. An MSA reveals extensive conservation of C66 (83%) and P67 (89%) in the primary sequence of DinB-like proteins, but not in all Y-family DNA polymerases (15% and 16%, respectively). (A) MSA of predicted and bona fide DinB-like sequences. (B) MSA of predicted and bona fide Y-family DNA polymerase sequences (DinB-like and UmuC-like). The dashes indicate a lack of consensus in the amino acid sequence. (C) DinBC66 and DinBP67 are surface residues, do not pertain to the active site, and are located at the same surface as F172. C66, P67, and F172 are highlighted in white. The in silico model of DinB was rendered using PyMol.
