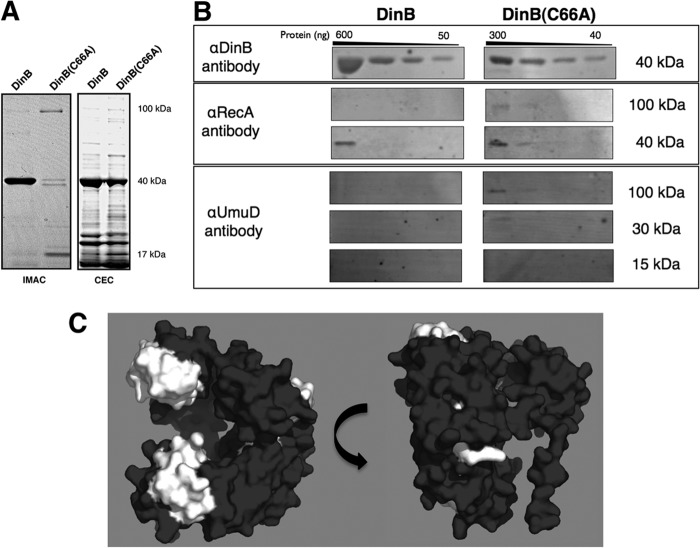
Fig 3.
The DinB(C66A) derivative copurifies with more RecA and UmuD than DinB. (A) SDS-PAGE of elution fractions for hexahistidine-tagged DinB and DinB(C66A) purified by IMAC (left) and native DinB and DinB(C66A) purified by CEC (right). The molecular masses shown are approximate. Seven micrograms of total protein was loaded for DinB-His purified by IMAC, and 1.3 μg of total protein was loaded for DinB(C66A)-His. For proteins purified by CEC, 6 μg of total protein was loaded for native DinB and 3 μg of total protein was loaded for native DinB(C66A). (B) Immunoblot of pooled elution fractions for DinB and DinB(C66A) purified by CEC. For all immunoblots, undiluted and serially diluted pooled elution fractions were loaded; 3 μg of total protein was loaded in the undiluted sample of DinB, while 1.5 μg of total protein was loaded in the undiluted sample of DinB(C66A). Less DinB(C66A) than DinB was present in the sample (300 ng versus 600 ng), consistent with less total protein loaded (approximately half); however, DinB(C66A) copurified with more RecA, UmuD, and intact putative MPC than DinB. The amounts of proteins per lane were determined by standard curves of blotted serially diluted proteins developed with the respective antibodies. The molecular masses shown are approximate. The amounts of copurified RecA and UmuD were determined to be less than 40 ng. (C) DinB epitope map for anti-DinB (αDinB) antibodies. Epitopes recognized by the affinity-purified αDinB antibody were determined by peptide array mapping, as indicated in Materials and Methods. Recognized epitopes were mapped onto an in silico model of DinB and are highlighted in white. The epitopes recognized by the polyclonal antibody localize to the exposed surfaces of DinB that are predicted to be bound by UmuD and RecA. The in silico DinB model was rendered using PyMol.