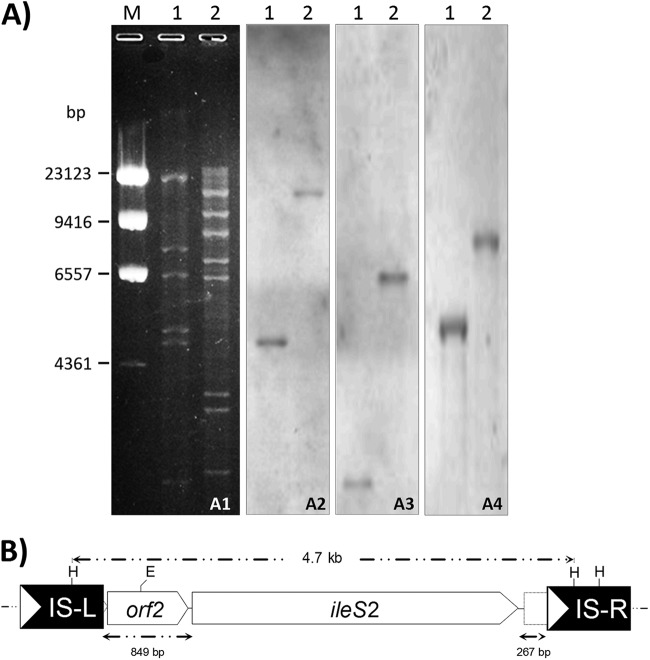
Fig 1.
(A) Analysis of plasmid pKM01 by restriction endonuclease analysis and Southern hybridization. (A1) Restriction endonuclease analysis of pKM01. Lane M, lambda phage DNA digested with HindIII as a molecular size standard; lane 1, pKM01 digested with HindIII; lane 2, pKM01 digested with EcoRI. Numbers on the left correspond to the molecular sizes (in base pairs) of λ DNA HindIII restriction fragments. Southern hybridization was carried out with the ileS2-containing DNA as a probe (A2), an aacA-aphD gene fragment as a DNA probe (A3), and a traK gene fragment as a DNA probe (A4). The lane contents of panels A2 to A4 are the same as in panel A1. (B) Genetic organization of IS257s flanking the ileS2 gene on the pKM01 plasmid, encoding Hi-Mupr. Restriction endonuclease cleavage sites are abbreviated as follows: E, EcoRI, and H, HindIII. Upstream (IS-L) and downstream (IS-R) IS257 elements flanking the ileS2 gene are represented by solid boxes; the white arrows indicate the direction of IS257 transposase transcription. The ileS2 gene and the predicted open reading frames (ORFs) up- and downstream are represented as arrows, with the arrowhead indicating their orientation. Truncated ORFs are shown using a dotted line. The broken arrows above the diagram note the extent of the HindIII fragment hybridizing with the ileS2 probe in restriction fragment length polymorphism (RFLP) analyses. Below are shown the sizes (in bp) of sequences corresponding to up- and downstream IS257-ileS2 spacers.