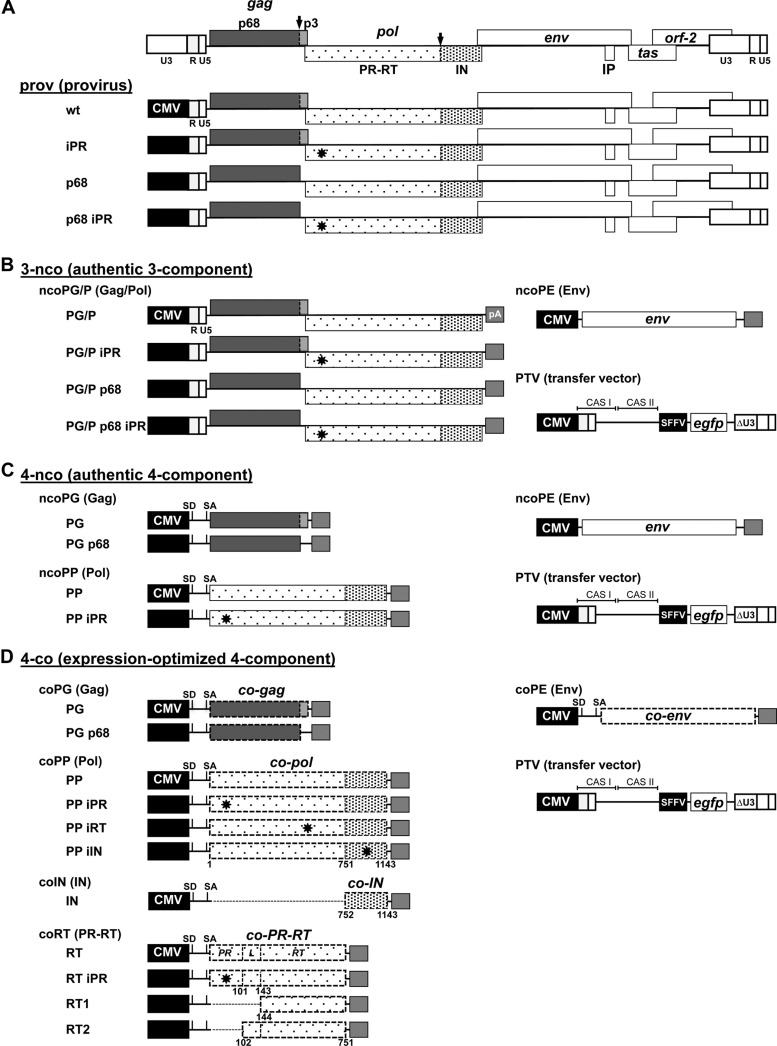
Fig 1.
PFV production systems. Schematic illustration of the different PFV production systems used in the study. Three systems based on authentic PFV ORFs were utilized and include systems for production of the following: prov, replication-competent virus based on CMV (cytomegalovirus)-driven proviral expression constructs (A); 3-nco, replication-deficient vectors based on CMV-driven Gag/Pol and Env packaging constructs and CMV-driven PFV transfer vector (PTV) containing an egfp marker gene expression cassette (B); and 4-nco, replication-deficient vectors based on separate CMV-driven packaging vectors for Gag, Pol, and Env and the previously mentioned transfer vector (C). (D) A fourth replication-deficient vector system (4-co) is based on separate packaging constructs with expression-optimized ORFs encoding PFV Gag, Pol, and Env and the same transfer vector mentioned above. R, long terminal repeat region (LTR); U5, LTR unique 5′ region; U3, LTR unique 3′ region; ΔU3: enhancer-promoter deleted U3 region; IP, internal promoter; CAS, cis-acting sequence; SFFV, spleen focus-forming virus U3 promoter; SD, splice donor; SA, splice acceptor; PR, protease domain; RT, reverse transcriptase domain; L, linker domain between PR and RT; IN: integrase domain; pA: bovine growth hormone polyadenylation site. Major PFV PR cleavage sites in PFV Gag and Pol are indicated by black arrows; iPR (D24A), iRT (DD312–315GAAA), and iIN (D936A) mutations are indicated with stars throughout the pol ORF; authentic ORFs are indicated by boxes with continuous lines; expression-optimized ORFs are indicated by boxes with dashed lines. Numbers indicate amino acid positions in Pol.