Fig 1.
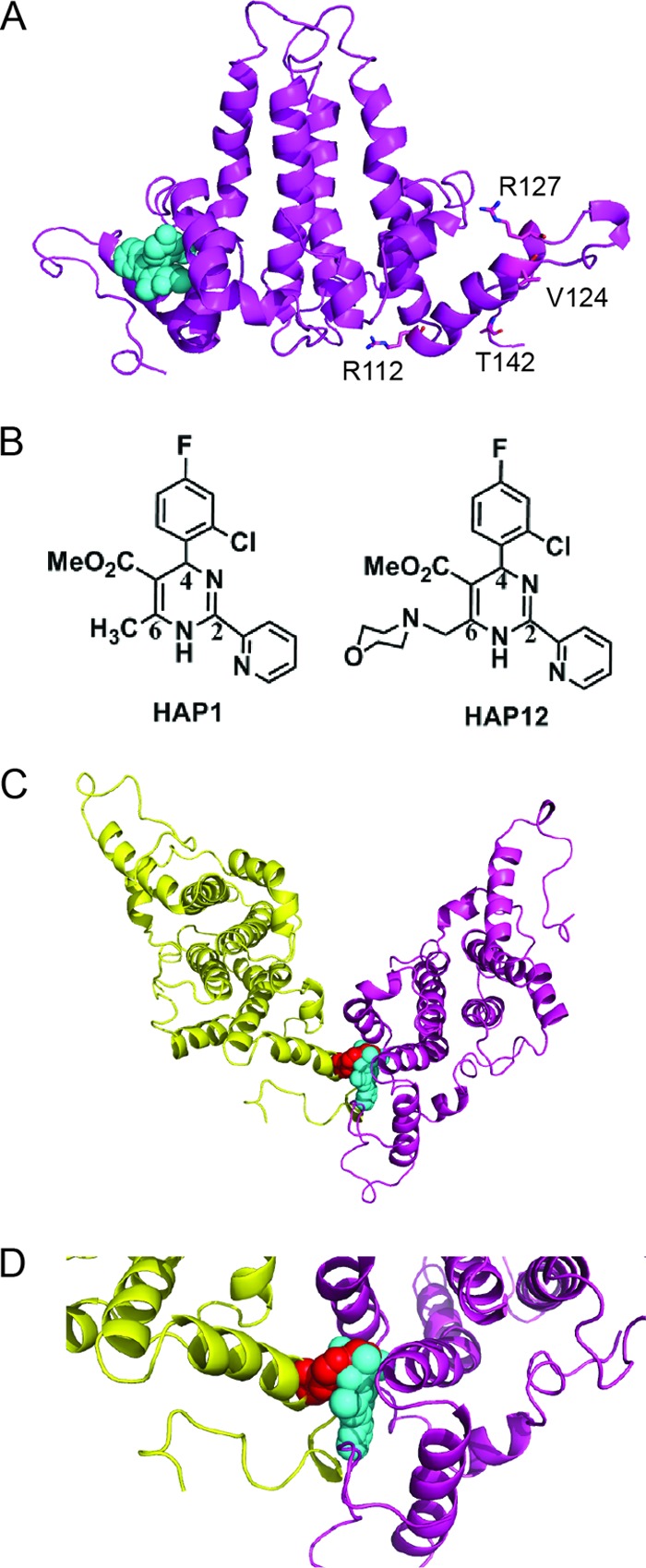
Model of Cp149-V124W dimer structure showing that the V124W mutant overlaps the HAP binding site. (A) Side view of the HBV Cp149-WT ribbon structure with bound HAP1 (cyan spheres) (PDB accession no. 2G34). Residues R112, R127, V124, and T142 are shown as sticks for reference points. (B) Structures of HAP1 and HAP12. HAP12 has an additional six-member ring linked to the methyl group at position 6. (C) Model of Cp149-V124W, generated based on Cp149-WT (PDB accession no. 2G34) with WinCoot by mutating V124 of the core protein D subunit to tryptophan. The selected rotamer accounts for 32% of the observed tryptophan rotamers. Viewed from the capsid interior, the HAP1 binding site is seen at the dimer-dimer interface. The V124W mutation (red spheres) overlaps HAP1 and partially fills the HAP binding site. The two dimers forming the site are colored yellow and magenta. (D) Close-up view of the HAP binding site at the V124W dimer-dimer interface.
