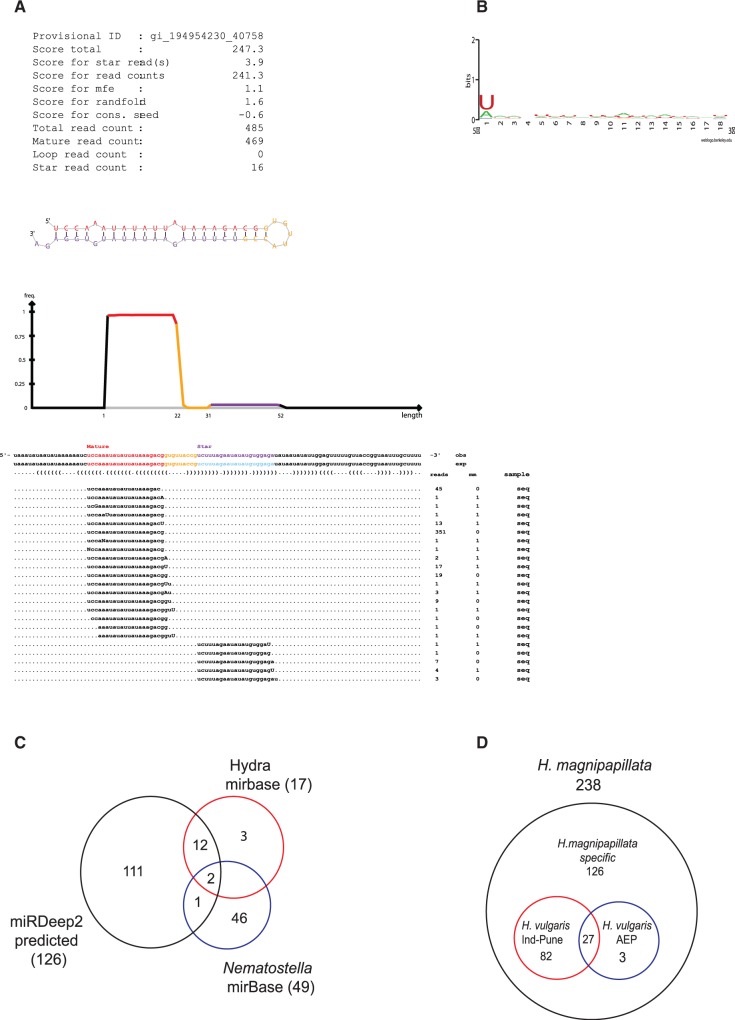
Figure 2.
Identification of hydra miRNAs using miRDeep2. (A) Figure shows sequence of a typical miRNA (hma-mir-new46) identified by miRDeep2, showing mature strand and star strand and the frequency of the reads obtained for both strands during deep sequencing. (B) Base preference for predicted miRNAs. All predicted miRNAs show preference for U at 5′-end indicating processing by dicer. (C) Pie chart depicting miRNAs that are common between set of miRNAs identified in this study, hydra miRNAs present in miRBase and N. vectensis miRNAs present in miRBase. (D) 18–24 nt reads from H. vulgaris Ind-Pune and H. vulgaris AEP were mapped to miRNAs identified in H. magnipapillata allowing no mismatch and figure shows miRNAs conserved across these species of hydra.